-
Notifications
You must be signed in to change notification settings - Fork 5
App ultraVessMorpho2Mesh
ultraVessMorpho2Mesh reconstructs high fidelity, multi-partitioned and two manifold watertight surface mesh models of vascular networks from corresponding fragmented and large-scale vascular graphs. Resulting meshes can be optimized to produce high quality topology with resonable tessellation as shown in Figure 1.
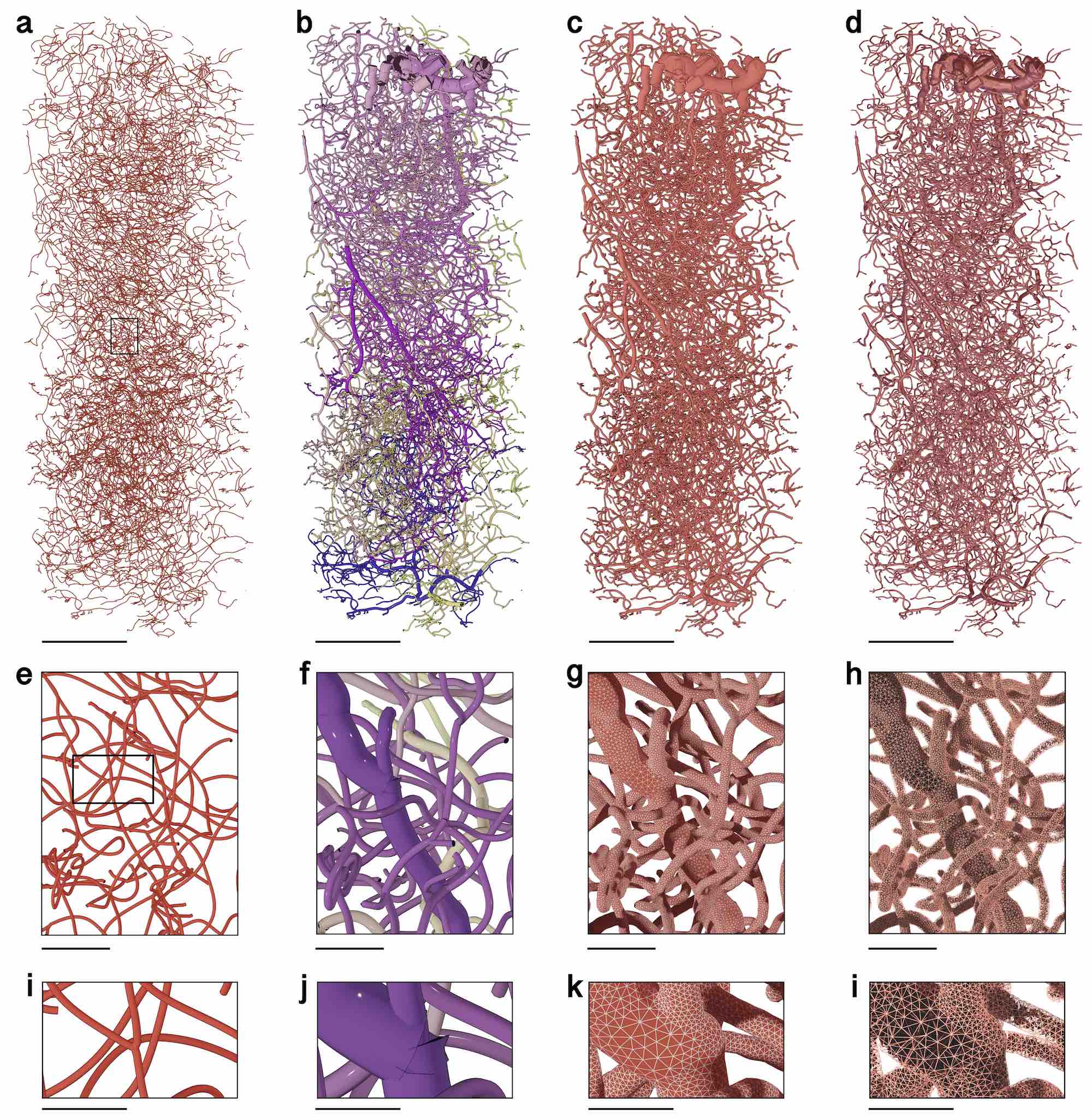
Figure 1
Vasculature Mesh Reconstruction. Steps showing the conversion of a large-scale, highly detailed vascular graph into a corresponding tetrahedral volumetric mesh tailored to perform blood transport simulations. (a) Thin-line representation of the input vascular graph showing its connectivity. (b) Polyline representation of the graph showing the actual radii at each vertex in the graph. (c) The polyline representation is converted with ultraVessMorpho2Mesh into a watertight manifold having multiple mesh partitions. (d) Reconstruction of a tetrahedral volumetric mesh from the watertight mesh reconstructed with Ultraliser. TetGen is used to reconstruct this tetrahedral mesh. The closeups in (e - l) focus on a branching point. (k) Wireframe rendering of the surface mesh showing the accurate and adaptive topology of the manifold. (l) Wireframe rendering of the tetrahedral mesh visualizing the internal structure of the volume. Scale bars, 200 microns (a-d), 25 microns (e-h), 10 microns (i-l).
skeletons that are either stored in .SWC or .H5 file formats. Somata profiles are reconstructed on a physically plausible basis using a finite-element method (FEM) [1].
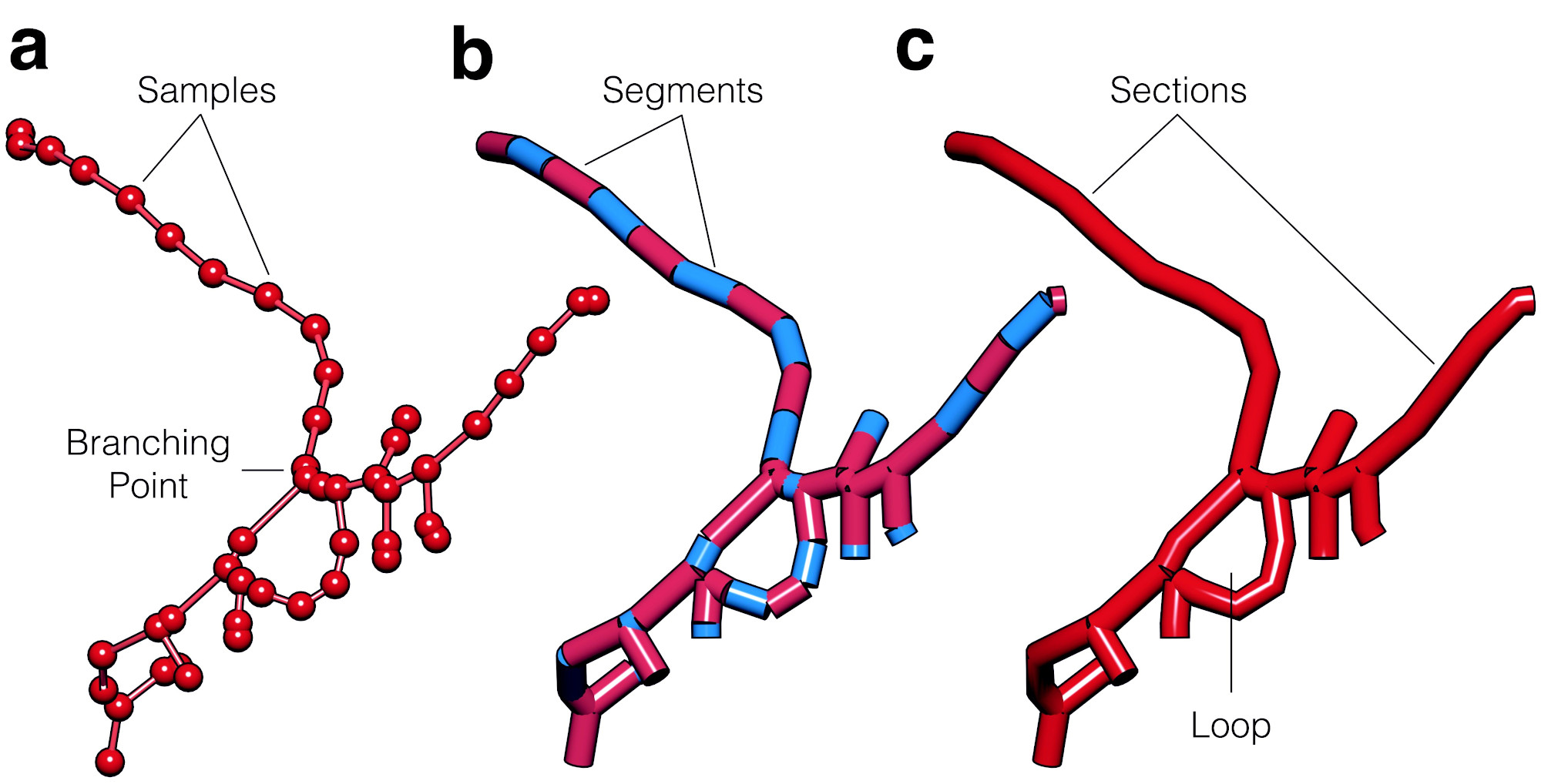
Figure 2: Vascular morphologies are represented with cyclic graphs to account for loops. The branching structure is similar to the arborizations of neurons and astrocytes. The skeleton is digitized into a list of connected samples (a), where every adjacent pair of samples represents a segment (b) and a group of connected segments between two branching points represents a section. Each section in the morphology has a unique index, and references to its parent and child sections, with which we can reconstruct the hierarchical organization of the graph. The loops are identified in the morphology when two sections have the same references to parent and child sections.
-
--morphologyThe absolute path of the input morphology. This application accepts only .SWC and .H5 astrocyte morphologies. For Neurolucida-based formats .ASC, please consult MorphIO to convert the .ASC morphologies to either .SWC or .H5 files.
-
--output-directoryThe absolute path of the parent directory where the results (or artifacts) will be generated. Resulting meshes will be created by default in themeshessubdirectory. If any of the projection flags are enabled, for example--project-xy, the resulting projection will be generated to theprojectionsdirectory. Further details on the structure of the output directory are available in this page. -
--prefixA file prefix that will be used to label the generated files. If this prefix is not given by the user, the base name of the input file will be considered to label all the output artifacts. For example, if the input morphology name isneuron1.h5, the resulting mesh should be labeledneuron1.obj. If a prefix is given, for example--prefix cortical_neuron, the resulting mesh will be labeledcortical_neuron.obj.
-
--resolutionThe base resolution of the volume that will be created to voxelize the input neuron morphology. This resolution is set to the larget dimension of the bounding box of the input morphology, while the resolutions of the other dimensions are computed accordingly. The default value is 512. -
--scaled-resolutionIf this flag is set, the resolution of the volume (that will be created to voxelize the input morphology) will be computed based on the dimensions of the morphology (in microns) and the--voxels-per-micronscale factor. For example, if the largest dimension of the input morphology is 100 microns, and the--voxels--per-micronsvalue is 5, the resolution of the volume will be 500. The value of the--resolutionargument is ignored if this flag is set. -
--voxels-per-micronNumber of voxels per micron needed to construct the volume grid that will be used to voxelize the input morphology. The value of this parameter is only considered if the--scaled--resolutionflag is set, otherwise the value defined by the--resolutionargument is used. The default value is 3.
--volume-type: Specify a volume format to perform the voxelization: [bit, byte, voxel]. By default, it is a bit volume to reduce the memory foot print.
-
--solidUse solid voxelization to fill the interior of the volume shell created from the rasterization (or surface voxelization) of the input morphology into the created volume grid. For this specific application, this option is advised TO BE ALWAYS SET. -
voxelization-axisThe axis where the solid voxelization operation will be performed. Use one of the following options [x, y, z, or xyz]. If you use x or y or z the voxelization will happen along a single axis, otherwise, using xyz will perform the solid voxelization along the three main axes of the volume to avoid filling any loops in the morphology. By default, the Z-axis solid voxelization is applied ONLY if the--solidflag is set. -
--bounds-fileA file that defines the bounding box or region of interest (ROI) that will be voxelized and meshed. This option is used to select a specifc ROI from the space to voxelize. This is a single-line ASCII file that contains pMin and pMax of the ROI in the following formatX_MIN Y_MIN Z_MIN X_MAX Y_MAX Z_MAX. -
--edge-gapSome little extra space (in percentage of the total bounding box the input astrocyte) to avoid edges intersection. The default value is 0.05.
-
--project-xyProject the volume along theZ-axisand create a gray-scale PPM image. -
--project-xzProject the volume along theY-axisand create a gray-scale PPM image. -
--project-zyProject the volume along theX-axisand create a gray-scale PPM image. -
--project-color-codedGenerate color-coded projections of the volume in different colormaps to help the investigation process. Further details on the colormaps are available in this page.
-
--export-stack-xyGenerate an image stack (volume slices) along theZ-axisof the volume. -
--export-stack-xzGenerate an image stack (volume slices) along theY-axisof the volume. -
--export-stack-zyGenerate an image stack (volume slices) along theX-axisof the volume.
-
--export-bit-volumeExport an Ultraliser-specific bit volume, where each voxel is stored in 1 bit. The header and data are stored in a single file with the extention .UVOL. Further details are available in this page. -
--export-unsigned-volumeExport an Ultraliser-specific unsigned volume, where each voxel is either in 1, 2, 3 or 4 bytes depending on the type of the volume. The header and data are stored in a single file with the extention .UVOL. Further details are available in this page. -
--export-raw-volumeExport a raw volume, where each voxel is stored in 1 byte. The resulting files are: .IMG file (contains data) and .HDR file (meta-data) -
--export-nrrd-volumeExport a .NRRD volume that is compatible with VTK and can be loaded with Paraview for visualization purposes. The resulting output contains the header and data integrated into a single .NRRD file.
-
--export-volume-meshExport a mesh that represents the volume where each voxel will be a cube. The format of the exported mesh(es) is specified by the Mesh Export Arguments -
--export-volume-bounding-box-mesh
Export a mesh that represents the bounding box of the volume. This mesh is primarily used for debugging purposes. The format of the exported mesh(es) is specified by the Mesh Export Arguments -
--export-volume-grid-meshExport a mesh that represents the volumetric grid used to voxelize the mesh. This mesh is primarily used for debugging purposes. The format of the exported mesh(es) is specified by the Mesh Export Arguments
-
--isosurface-techniqueSpecify a technique to extract the isosurface from the volume. The options are [mc, dmc] for marching cubes and dual marching cubes respectively. The default technique is dmc (Dual Marching Cubes). -
--preserve-partitionsKeeps all the partitions (islands) of the mesh if the resulting mesh contains more than one. For this specific application, this option is advised NOT TO BE SET.
-
--laplacian-iterationsNumber of iterations to smooth the reconstructed mesh with Laplacian filter. The default value is 10. -
--optimize-meshOptimize the reconstructed mesh using the default optimization strategy. -
--adaptive-optimizationOptimize the reconstructed mesh using the adaptive optimization strategy. -
--optimization-iterationsNumber of iterations to optimize the resulting mesh. The default value is 1. NOTE: If this value is set to 0, the optimization process will be ignored even if the--optimize-meshflag is set. -
--smooth-iterationsNumber of iterations to smooth the reconstructed mesh, The default value is 5. -
--flat-factorA factor that is used for the coarseFlat function. The default value is 0.05. -
--dense-factorA factor that is used for the coarseDense function. The default value is 5.0. -
--min-dihedral-angleThe required minimum dihedral angle. The default value is 0.1.
-
--x-scaleScaling factor for the mesh along the X-axis, The default value is 1.0. -
--y-scaleScaling factor for the mesh along the Y-axis. The default value is 1.0. -
--z-scaleScaling factor for the mesh along the Z-axis. The default value is 1.0.
-
--export-obj-meshExport the resulting mesh(es) to Wavefront object format (.OBJ). -
--export-ply-meshExport the resulting mesh(es) to the Stanford triangle format (.PLY). -
--export-off-meshExport the resulting mesh(es) to the object file format (.OFF). -
--export-stl-meshExport the resulting mesh(es) to the stereolithography CAD format (.STL). -
--ignore-marching-cubes-meshIf this flag is set, the mesh reconstructed with the marching cubes algorithm will be ignored and will NOT be written to disk. Setting this flag speeds up the process to create the final watertight mesh. Further details are available at this page. -
--ignore-laplacian-meshIf this flag is set, the mesh resulting from the application of the Laplacian operator will be ignored and will NOT be written to disk. Setting this flag speeds up the process to create the final watertight mesh. Further details are available at this page. -
--ignore-optimized-meshIf this flag is set, the optimized mesh will NOT be written to disk. Setting this flag speeds up the process to create the final watertight mesh. Further details are available at this page. -
--export-at-originExport the astrocyte mesh at the origin, where the center of the bounding box will be located at the origin O(0, 0, 0).
-
--statsWrite the statistics of the input and resulting meshes/volumes/morphologies. Further details are available in this page. -
--distsWrite the statistical distributions of the input and resulting meshes/volumes/morphologies. Further details are available in this page.
-
--threadsNumber of threads used to process the parallel chunks in the code. If this value is set to 0, all the cores available in the system will be used. The default value is 0.
In this page, we provide a list of examples to demonstrate how to use ultraVessMorpho2Mesh.