-
Notifications
You must be signed in to change notification settings - Fork 5
App ultraMesh2Mesh
ultraMesh2Mesh generates watertight surface mesh models from non-watertight polygonal soups. The resulting mesh can be optimized to produce a smooth and continuous surface with high fidelity topology and resonable tessellation as shown in Figure 1.
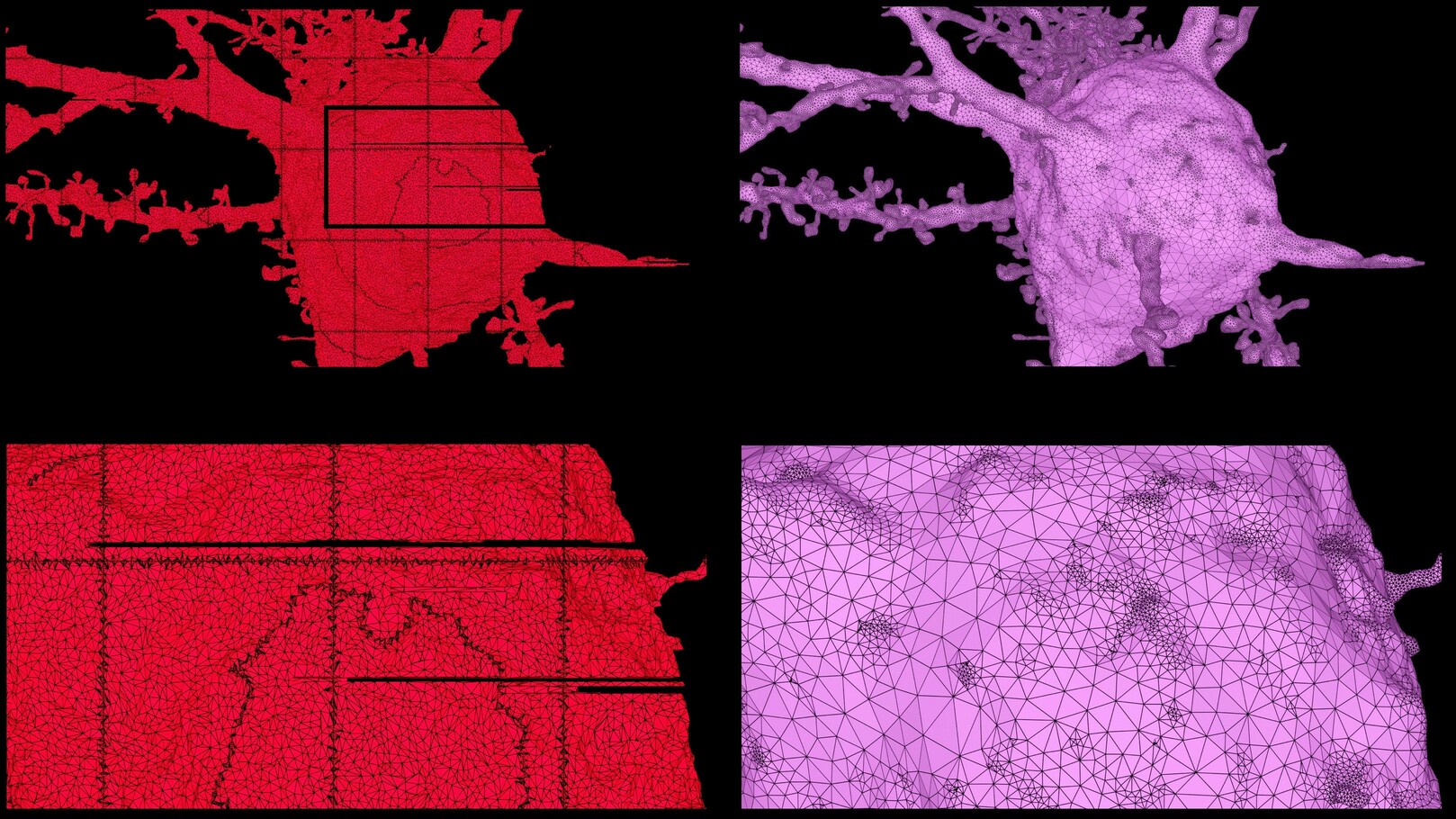
Figure 1 Reconstruction of high fidelity surface mesh (right) from an input triangular soup (left). The input mesh represents a pyramidal neuron segmented from layer 2/3 of the mouse visual cortex1, 2, 3 and is available from a recent dataset by the MICrONS Explorer.
The input triangular soup is rasterized into a binary volume grid, where each voxel is represented by a single bit. After the ratserization, the interior of the grid is filled based on slice-based solid voxelization. The final volume grid is used to reconstruct a coarse mesh based on either marching cubes or dual marching cubes. A Laplacian filter is applied to this coarse mesh to smooth its surface. The smoothed mesh is then optimized (or adaptively optimized) to create a high-fidelity mesh with super nice topology and convenient tessellation level. This optimization process might create some self-intersecting triangles along the surface of the mesh depending on its complexity. However, and in a subsequent final stage, the self-intersections are detected and removed to create a two-manifold watertight mesh with zero non-manifold edges and non-manifold vertices and no self-intersecting triangles, i.e. a watertight mesh.
-
--meshThe absolute path to the input mesh. Supported mesh types: .OBJ, .PLY, .STL, .OFF and .H5. The format of the H5 files is specified by the MICrONS Explorer (microns-explorer.org). If you wish to integrate your specific format, please contact the corresponding authors.
-
--output-directoryThe absolute path of the parent directory where the results (or artefacts) will be generated. Resulting meshes will be created by default in themeshessubdirectory. If any of the projection flags are enabled, for example--project-xy, the resulting projection will be generated to theprojectionsdirectory. Further details on the structure of the output directory are available in this page. -
--prefixA file prefix that will be used to label the generated files. If this prefix is not given by the user, the base name of the input file will be considered to label all the output artefacts. For example, if the input mesh name isneuron-1.obj, the resulting mesh will be labeledneuron-1-*.obj. If a prefix is given, for example--prefix cortical_neuron, the resulting mesh will be labeledcortical_neuron-*.obj.
-
--resolutionThe base resolution of the volume that will be created to voxelize the input mesh. This resolution is set to the largest dimension of the bounding box of the input mesh, while the resolutions of the other dimensions are computed accordingly. The default value is 512. -
--scaled-resolutionIf this flag is set, the resolution of the volume (that will be created to voxelize the input mesh) will be computed based on the dimensions of the mesh (in microns) and the--voxels-per-micronscale factor. For example, if the largest dimension of the input mesh is 100 microns, and the--voxels--per-micronsvalue is 5, the resolution of the volume will be 500. The value of the--resolutionargument is ignored if this flag is set. -
--voxels-per-micronNumber of voxels per micron needed to construct the volume grid that will be used to voxelize the input mesh. The value of this parameter is only considered if the--scaled--resolutionflag is set, otherwise the value defined by the--resolutionargument is used. The default value is 3. -
--solidUse solid voxelization to fill the interior of the volume shell created from the rasterization (or surface voxelization) of the input mesh into the created volume grid. -
voxelization-axisThe axis where the solid voxelization operation will be performed. Use one of the following options [x, y, z, or xyz]. If you use x or y or z, the voxelization will happen along a single axis, otherwise, using xyz will perform the solid voxelization along the three main axes of the volume to avoid filling any loops in the morphology. By default, the Z-axis solid voxelization is applied ONLY if the--solidflag is set. -
--bounds-fileA file that defines the bounding box or region of interest (ROI) that will be voxelized and meshed. This option is used to select a specifc ROI from the space to voxelize. This is a single-line ASCII file that contains pMin and pMax of the ROI in the following formatX_MIN Y_MIN Z_MIN X_MAX Y_MAX Z_MAX. -
--edge-gapSome little extra space (in percentage of the total bounding box the input astrocyte) to avoid edges intersection. The default value is 0.05.
-
--project-xyProject the volume along theZ-axisand create a gray-scale PPM image. -
--project-xzProject the volume along theY-axisand create a gray-scale PPM image. -
--project-zyProject the volume along theX-axisand create a gray-scale PPM image. -
--project-color-codedGenerate color-coded projections of the volume in different colormaps to help the investigation process. Further details on the colormaps are available in this page.
-
--export-stack-xyGenerate an image stack (volume slices) along theZ-axisof the volume. -
--export-stack-xzGenerate an image stack (volume slices) along theY-axisof the volume. -
--export-stack-zyGenerate an image stack (volume slices) along theX-axisof the volume.
-
--export-bit-volumeExport an Ultraliser-specific bit volume, where each voxel is stored in 1 bit. The header and data are stored in a single file with the extention .UVOL. Further details are available in this page. -
--export-unsigned-volumeExport an Ultraliser-specific unsigned volume, where each voxel is either in 1, 2, 3 or 4 bytes depending on the type of the volume. The header and data are stored in a single file with the extention .UVOL. Further details are available in this page. -
--export-raw-volumeExport a raw volume, where each voxel is stored in 1 byte. The resulting files are: .IMG file (contains data) and .HDR file (meta-data) -
--export-nrrd-volumeExport a .NRRD volume that is compatible with VTK and can be loaded with Paraview for visualization purposes. The resulting output contains the header and data integrated into a single .NRRD file.
-
--export-volume-meshExport a mesh that represents the volume where each voxel will be a cube. The format of the exported mesh(es) is specified by the Mesh Export Arguments -
--export-volume-bounding-box-mesh
Export a mesh that represents the bounding box of the volume. This mesh is primarily used for debugging purposes. The format of the exported mesh(es) is specified by the Mesh Export Arguments -
--export-volume-grid-meshExport a mesh that represents the volumetric grid used to voxelize the mesh. This mesh is primarily used for debugging purposes. The format of the exported mesh(es) is specified by the Mesh Export Arguments
-
--isosurface-techniqueSpecify a technique to extract the isosurface from the volume. The options are [mc, dmc] for marching cubes and dual marching cubes respectively. The default technique is dmc (Dual Marching Cubes). -
--preserve-partitionsKeeps all the partitions (islands) of the mesh if the resulting mesh contains more than one. For this specific application, this option is advised NOT TO BE SET.
-
--laplacian-iterationsNumber of iterations to smooth the reconstructed mesh with Laplacian filter. The default value is 10. -
--optimize-meshOptimize the reconstructed mesh using the default optimization strategy. -
--adaptive-optimizationOptimize the reconstructed mesh using the adaptive optimization strategy. -
--optimization-iterationsNumber of iterations to optimize the resulting mesh. The default value is 1. NOTE: If this value is set to 0, the optimization process will be ignored even if the--optimize-meshflag is set. -
--smooth-iterationsNumber of iterations to smooth the reconstructed mesh, The default value is 5. -
--flat-factorA factor that is used for the coarseFlat function. The default value is 0.05. -
--dense-factorA factor that is used for the coarseDense function. The default value is 5.0. -
--min-dihedral-angleThe required minimum dihedral angle. The default value is 0.1.
-
--x-scaleScaling factor for the mesh along the X-axis, The default value is 1.0. -
--y-scaleScaling factor for the mesh along the Y-axis. The default value is 1.0. -
--z-scaleScaling factor for the mesh along the Z-axis. The default value is 1.0.
-
--export-obj-meshExport the resulting mesh(es) to Wavefront object format (.OBJ). -
--export-ply-meshExport the resulting mesh(es) to the Stanford triangle format (.PLY). -
--export-off-meshExport the resulting mesh(es) to the object file format (.OFF). -
--export-stl-meshExport the resulting mesh(es) to the stereolithography CAD format (.STL). -
--ignore-marching-cubes-meshIf this flag is set, the mesh reconstructed with the marching cubes algorithm will be ignored and will NOT be written to disk. Setting this flag speeds up the process to create the final watertight mesh. Further details are available at this page. -
--ignore-laplacian-meshIf this flag is set, the mesh resulting from the application of the Laplacian operator will be ignored and will NOT be written to disk. Setting this flag speeds up the process to create the final watertight mesh. Further details are available at this page. -
--ignore-optimized-meshIf this flag is set, the optimized mesh will NOT be written to disk. Setting this flag speeds up the process to create the final watertight mesh. Further details are available at this page. -
--export-at-originExport the astrocyte mesh at the origin, where the center of the bounding box will be located at the origin O(0, 0, 0).
-
--statsWrite the statistics of the input and resulting meshes/volumes/morphologies. Further details are available in this page. -
--distsWrite the statistical distributions of the input and resulting meshes/volumes/morphologies. Further details are available in this page.
-
--threadsNumber of threads used to process the parallel chunks in the code. If this value is set to 0, all the cores available in the system will be used. The default value is 0.
In this page, we provide a list of examples to demonstrate how to use ultraMesh2Mesh.
-
Binary and analog variation of synapses between cortical pyramidal neurons. Dorkenwald, S., Turner, N.L., Macrina, T., Lee, K., Lu, R., Wu, J., Bodor, A.L., Bleckert, A.A., Brittain, D., Kemnitz, N., et al. (2019). bioRxiv 2019.12.29.890319; doi: https://doi.org/10.1101/2019.12.29.890319
-
Structure and function of axo-axonic inhibition Schneider-Mizell, C. Bodor, A.L., Collman, F. Brittain,D. Bleckert, AA, Dorkenwald, S., Turner N.L. Macrina, T. Lee, K. Lu, R. Wu, J. et al. eLife 2021;10:e73783 DOI: 10.7554/eLife.73783
-
Reconstruction of neocortex: Organelles, compartments, cells, circuits, and activity Nicholas L. Turner*, Thomas Macrina*, J. Alexander Bae*, Runzhe Yang*, Alyssa M. Wilson*, Casey Schneider-Mizell*, Kisuk Lee1*, Ran Lu*, Jingpeng Wu*, Agnes L. Bodor*, Adam A. Bleckert*, Derrick Brittain*, Emmanouil Froudarakis*, Sven Dorkenwald*, Forrest Collman*, Nico Kemnitz* et al. Cell, Volume 185, Issue 6, 2022, Pages 1082-1100.e24, https://doi.org/10.1016/j.cell.2022.01.023.
-
MICrONS Explorer: A virtual observatory of the cortex. Seung Lab - Princeton University, Brain-map.org - Allen Institute for Brain Science, Tolias Lab - Baylor College of Medicine and IARPA Microns.