-
Notifications
You must be signed in to change notification settings - Fork 8
Pathway analysis
apulvirenti edited this page Apr 23, 2021
·
10 revisions

Once the differential expression analysis is completed (for the guideline please refer to the previous section), it is also possible to perform the pathway analysis. It is important to clarify that the pathway analysis can only be performed on mRNAs and miRNAs, which have been found differentially expressed by the previous analysis. To start the pathway analysis, click Run Analysis on the dashboard and then New Pathway Analysis. After that, follow the step-by-step instructions described through the user interface to set and start the analysis. For pathway analysis, three steps are required.
- Choose name and type. Here users can write the sample code, the analysis name, and selected the results of the differential expression analysis to be used for the pathway analysis (All the completed differential expression analysis are automatically listed in the Select one DEGs analysis bar). Click Next to proceed with the next step.

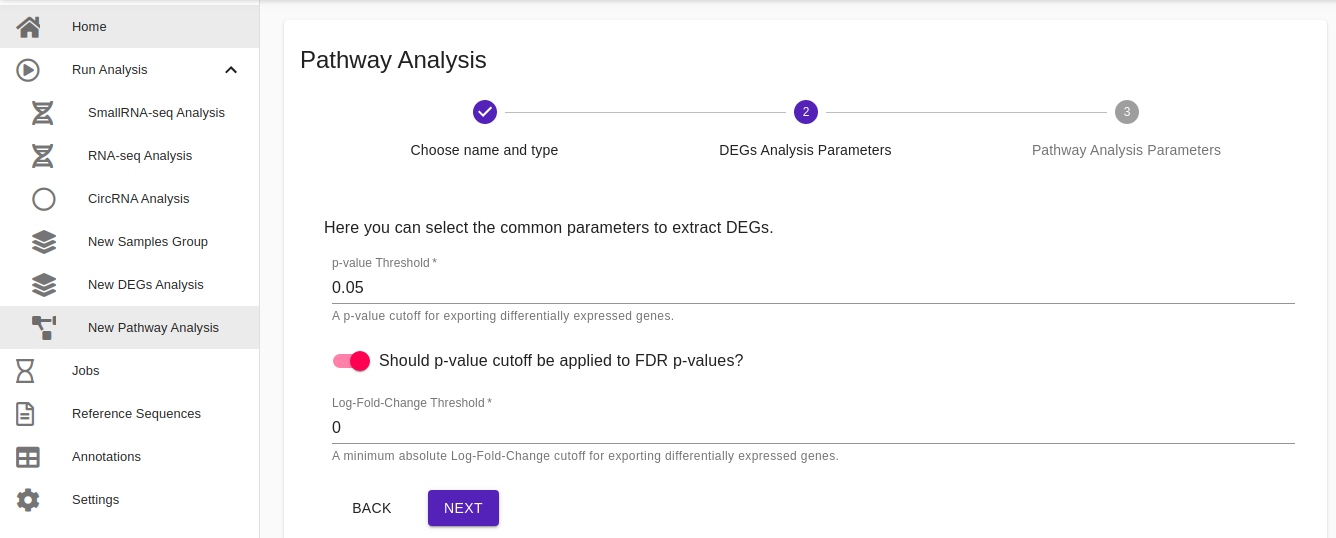
- DEGs Analysis Parameters. Here users can choose the criteria for the selection of the differentially expressed transcripts (mRNAs and miRNAs) to be used for the pathway analysis by writing the p-value cutoff and the Log-Fold change threshold. Click Next to proceed with the next step.
- Pathway Analysis Parameters. Here users can choose the organism to be used for the pathway analysis and the p-values threshold for the selection of the statistically significant perturbated pathways. After entering all the parameters, users can start the analysis by clicking on the Start Analysis button.
