-
Notifications
You must be signed in to change notification settings - Fork 8
Add reference and annotation files

Before proceeding with the analysis, users have to download reference genomes and transcriptomes from our remote repository and available through the Reference Sequences section (see the example figures below).

To download reference genomes or transcriptomes from our remote repository click on the icon Install from repository that appears after few seconds in the upper right section of the user interface. Notice that the icon will not show if no internet connection is available. Next, select one or more genomes/transcriptomes to download and click Install to begin the process.

As soon as you click the Install button, a new job will be created. You can follow its progress in the Jobs section. Once the job finishes, all selected genomes/transcriptomes will appear in the references list. All genome annotations included in the selected genomes/transcriptomes will also appear in the Annotations list.
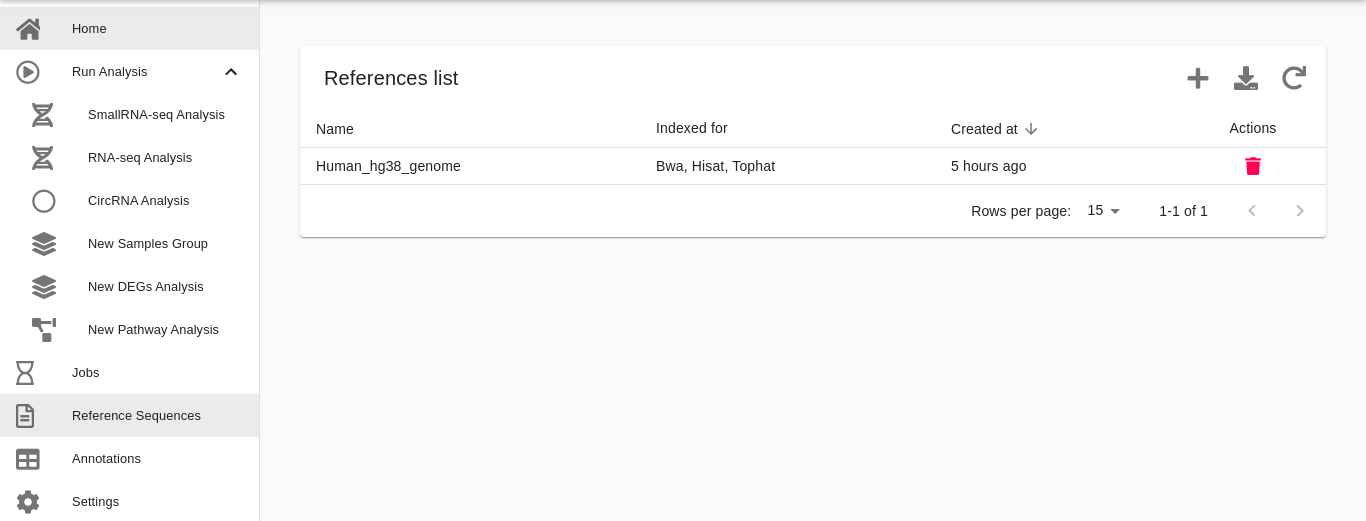
On the other hand, users can upload additional genomes/transcriptomes from their local computers by using the Reference Sequences section available from our dashboard. The user interface has a step-by-step procedure to perform such upload (please refer to the next paragraph).
If users want to analyze additional species, they can do it by uploading their genomes/transcriptomes in FASTA format. FASTA genomes of many organisms can be download from ENSEMBL or UCSC Genome Browser databases. FASTA genomes \ transcriptome can then be uploaded from the Reference Sequences section of our dashboard by clicking the icon Add in the upper right section of the user interface and follow the step-by-step procedure detailed in the user interface (see the example figures below).

- Choose a name. Write the name of the reference organism. It must contain only letters, numbers, and dashes.
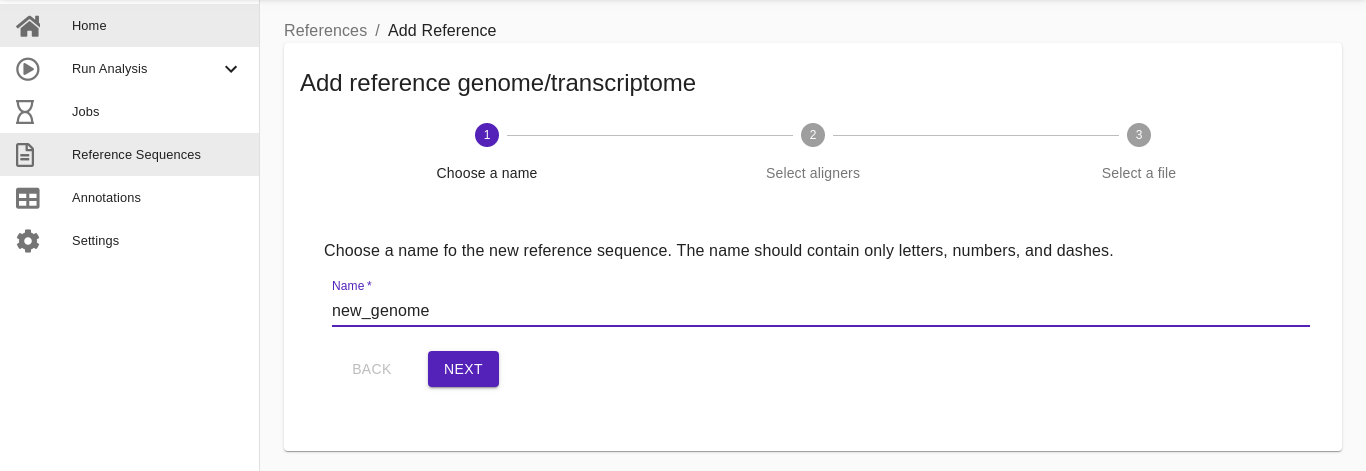
- Select aligners. Choose which alignment tool will be used for these sequences (users can select more than one). Based on the selected algorithms (BWA, STAR, HISAT2, SALMON), the appropriate indexed methods will be used.
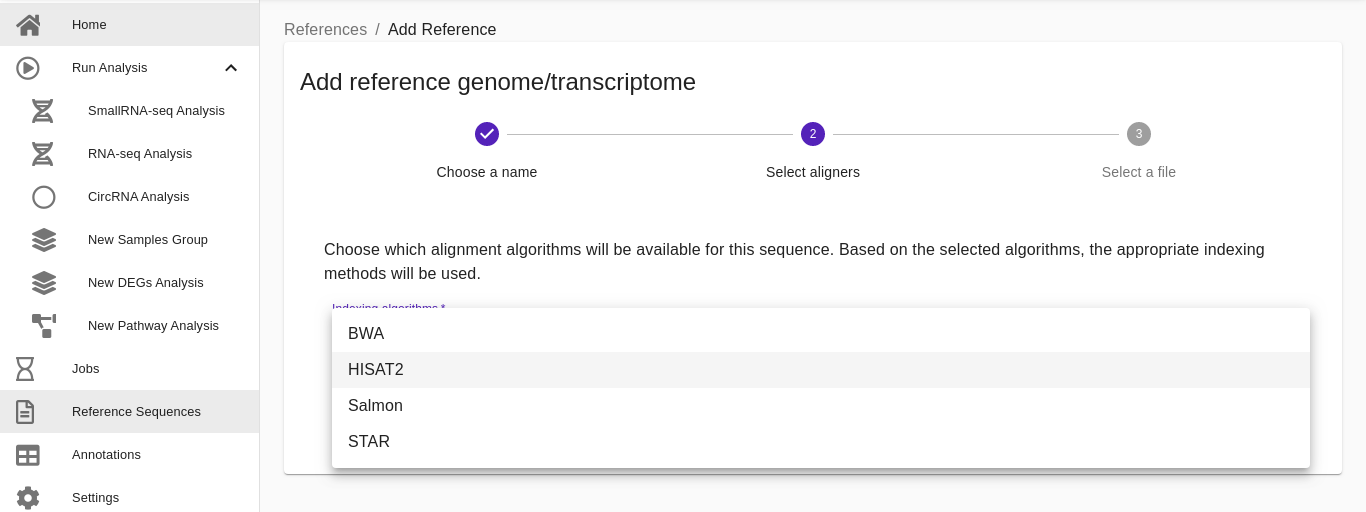
Note: to index medium\large genomes more than 32GB of RAM might be required. If you are planning to analyze a custom medium\large genome, you should consider installing RNAdetector in a local powerful server or in a cloud environment. Please consult the following reference here for more details concerning the installation of RNAdetector on one of the supported cloud environments.
- Select a file. Upload the genome \ transcriptome sequence in FASTA format and click Save to start the upload and index process.

If users want to analyze additional ncRNAs classes or mRNAs of additional species (read the previous paragraph to see how to analyze additional species), they can do it by uploading their genomic coordinates in GTF or BED format. Additional GTF or BED genome annotation files can be uploaded from the Annotations section of our dashboard by clicking the icon Add in the upper right section of the user interface and follow the step-by-step procedure detailed in the user interface (see the example figures below).
Note: The GTF files of all organisms whose genome has been sequenced can be found on Ensembl. Other databases can also be used to download the GTF files of specific classes of ncRNAs.

- Choose a name. Write a name for the new annotation file. It has to contain only letters, numbers, and dashes.

- Select a type. Select the type of the annotation file (GTF or BED).

- Select a file. Upload the annotation file and click Save to start the upload process.
