Objective: Model the incorporation of the FeGP cofactor into the hydrogenase and its catalytic capabilities. Context: Fe-hydrogenase is the third type of hydrogenase which requires a cofactor, the FeGP cofactor. The kinetics of incorporation, activation, and catalysis aren't yet fully understood. This work was done as part of the following study: Crystal Structures of [Fe]-Hydrogenase from Methanolacinia paynteri Suggest a Path of the FeGP-Cofactor Incorporation Process
Significance: Understanding cofactor binding and enzyme activation kinetics is an ongoing problem in biochemistry, with various approaches being developed. Mathematical modeling and computational simulation are among these approaches, applied here for the first time to the anaerobic Fe-hydrogenase.
Goal: Determine if observed kinetics can be explained through a mathematical model which further expands the reconstitution hypothesis.
- Gangfeng Huang
- Francisco J. Arriaza G.
- Tristan Wagner
- Seigo Shima
The project consists of several Python files for each condition studied. The kinetic approximation and ODEs derivation can be found in /plots/ODEs_S7 and ODEs_S9, as stated in the supplementary figures of the publication. One master script with the corresponding ODEs is found under /models/hcr.py
Experimental kinetic data performed at the Microbial Protein Structure group led by Dr. Seigo Shima at the Max Planck Institute for Terrestrial Microbiology.
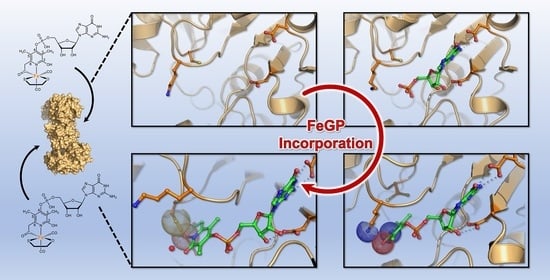
Structural studies of the [Fe]-hydrogenase in M. payntheri revealed conformational changes in several stages of the binding of the FeGP cofactor. In this trajectory, the residue Lys150 showed a significant difference between the unbound and bound states. To test the importance of this residue and its impact on the kinetics of binding a series of kinetic models were developed. First, the kinetic parameters of the mutants were determined by simulating the reaction progression curves as follow:
 Apparent Km and Vm were calculated by assuming an excess of H2 and/or H+ and approximated to a Michaelis-Menten equation.
Apparent Km and Vm were calculated by assuming an excess of H2 and/or H+ and approximated to a Michaelis-Menten equation.
 Increase of the Km values of the Lys150Ala variant is consistent with the observation of the contact of Lys150 with the side chain of the substrate described in sections 2.1-2.3 of the published work.
Increase of the Km values of the Lys150Ala variant is consistent with the observation of the contact of Lys150 with the side chain of the substrate described in sections 2.1-2.3 of the published work.
To determine the trajectory of binding and any impact of the mutation of Lys150, the binding and reactions were simulated together using the following model:
We calculated the specific activity (U/mg) at each time point (per 2 s) from the slope of the absorbance change. In these experiments, we assume that the increase of the Hmd activity indicates the increase of the active holoenzyme in the assay by incorporation of the FeGP cofactor into the apoenzymes. Hence, the specific activity is a function of the concentration of the reconstituted enzyme and the residual substrate concentration in the assay.
In the assay condition at pH 7.5, which is the physiological pH, the wild-type enzyme was quickly reconstituted and reached maximum activity within 25s in the presence of 700-nM FeGP cofactor (Subplot c). The Lys150Ala variant also exhibits activity, but the increase of the specific activity at the same condition was much slower than the case of the wild-type enzyme; to reach the maximal activity, 130s was required (Subplot d). These results support the hypothesis that Lys150 contributes to the binding kinetics of the FeGP cofactor to the protein.
Crystal Structures of [Fe]-Hydrogenase from Methanolacinia paynteri Suggest a Path of the FeGP-Cofactor Incorporation Process.
Gangfeng Huang, Francisco Javier Arriaza-Gallardo, Tristan Wagner and Seigo Shima