-
Notifications
You must be signed in to change notification settings - Fork 18
HarryPlotter
This tutorial/documentation is superseded by Artus/wiki/HarryPlotter-Overview.
This tutorial uses the outputs of the synchonisation analysis as described in Running Artus. The ROOT output file is denoted by output.root. It can be the output of an individual run, the one of an individual job or the merged output file.
HarryPlotter is part of the Artus repository. To only check out HarryPlotter, do the following:
git clone --branch dictchanges_CMSSW102 [email protected]:artus-analysis/Artus.git -n
cd Artus
git remote add -f Artus [email protected]:artus-analysis/Artus.git
git config core.sparsecheckout true
echo "/HarryPlotter/" >> .git/info/sparse-checkout
echo "/Utility/python/" >> .git/info/sparse-checkout
echo "/Utility/scripts/" >> .git/info/sparse-checkout
git pull
git read-tree --empty
git read-tree -mu HEADThe Artus run consisted of several pipelines. Each pipeline creates a separated TDirectory in the ROOT file and can be identified by looking into the file or the configuration.
artusPipelines.py output.rootThis should produce an output similar to
em
et
mt
tt
Each pipeline writes several objects into this directory:
get_root_file_content.py output.rootgives something similar to
config (TObjString)
em/ntuple (TTree)
em/cutFlowUnweighted (TH1F)
em/cutFlowWeighted (TH1F)
et/ntuple (TTree)
et/cutFlowUnweighted (TH1F)
et/cutFlowWeighted (TH1F)
mt/ntuple (TTree)
mt/cutFlowUnweighted (TH1F)
mt/cutFlowWeighted (TH1F)
tt/ntuple (TTree)
tt/cutFlowUnweighted (TH1F)
tt/cutFlowWeighted (TH1F)
...
HarryPlotter provides the executable harry.py. This script is extended with analysis-specific needs and then called higgsplot.py. Basic functionallity is possible with harry.py [-h], but here it is always referred to higgsplot.py [-h].
HarryPlotter can read in ROOT histograms, graphs and similar objects as well as create histograms or graphs from trees. Possible quantities to plot can be obtained with the following command:
higgsplot.py -i <input> -f <folder> --quantities
Based on the example, one could do
higgsplot.py -i output.root -f mt/ntuple --quantities
higgsplot.py -i output.root -f mt --quantities
which then gives
List of all tree quantities (in the first file):
eta_1 (Float_t)
eta_2 (Float_t)
m_vis (Float_t)
pt_1 (Float_t)
pt_2 (Float_t)
weight (Float_t)
...
and
List of all histogram/graph/function quantities (in the first file):
cutFlowUnweighted (TH1F)
cutFlowWeighted (TH1F)
This works for every ROOT file and is not limited to Artus outputs. HarryPlotter can plot contents of arbitrary ROOT files.
higgsplot.py -i output.root -f mt/ntuple -x m_vishiggsplot.py -i output.root -f mt/ntuple -x m_vis -w weight
higgsplot.py -i output.root -f mt/ntuple -x m_vis -w "weight*(eta_1>0)"higgsplot.py -i output.root -f mt/ntuple -x m_vis --x-bins "15,20,170"
higgsplot.py -i output.root -f mt/ntuple -x m_vis --x-bins "15 30 45 60 75 90 105 120 170 220 270"higgsplot.py -i output.root -f mt/ntuple et/ntuple -x m_vis [--labels MT ET --legend 0.5 0.6 0.8 0.8]
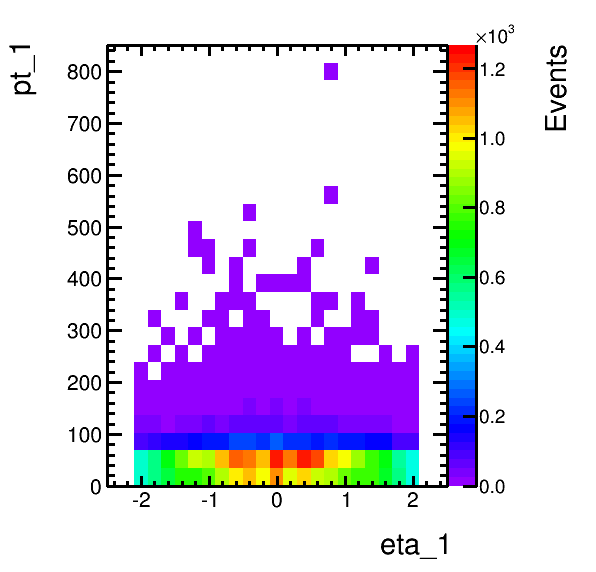
higgsplot.py -i output.root -f mt/ntuple -x m_vis "m_vis+10" [--labels m "m+10" --legend 0.5 0.6 0.8 0.8]higgsplot.py -i output.root -f mt/ntuple -x eta_1 -y pt_1 -m COLZhiggsplot.py -i output.root -f mt/ntuple -x eta_1 -y pt_1 --tree-draw-options prof -m Ehiggsplot.py -i output.root -f mt -x cutFlowWeightedhiggsplot.py --list-available-modules
higgsplot.py --analysis-modules <modules> -hhiggsplot.py -i output.root -f mt/ntuple -x m_vis --analysis-modules NormalizeToUnityhiggsplot.py -i output.root -f mt/ntuple et/ntuple -x m_vis --analysis-modules Ratio
higgsplot.py -i output.root -f mt/ntuple et/ntuple -x m_vis --analysis-modules Ratio --nicks mt et --ratio-numerator-nicks mt --ratio-denominator-nicks et --ratio-result-nicks ratioThese two commands are equivalent.

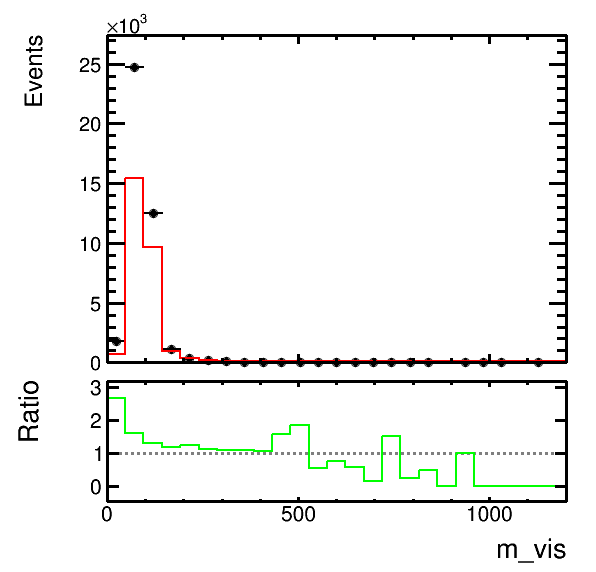
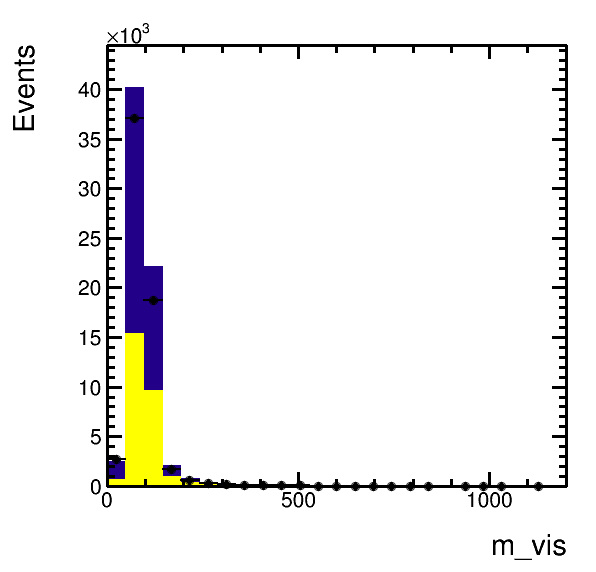
higgsplot.py -i output.root -f mt/ntuple et/ntuple mt/ntuple -x m_vis -w weight weight "1.5*weight" --stacks mc mc data -m HIST HIST E --colors kGreen kRed kBlack
higgsplot.py -i output.root -f mt/ntuple et/ntuple mt/ntuple -x m_vis -w weight weight "1.5*weight" --stacks mc mc data -m HIST HIST E E --colors kGreen kRed kBlack kBlack --nicks mc1 mc2 data --analysis-modules Ratio --ratio-numerator-nicks data --ratio-denominator-nicks "mc1 mc2"higgsplot.py -i output.root -f "mt/ntuple et/ntuple" -x m_vis
higgsplot.py -i output.root -f mt/ntuple et/ntuple -x m_vis --nicks mc mc
higgsplot.py -i output.root -f mt/ntuple et/ntuple -x m_vis --nicks mt_noplot et_noplot --analysis-modules AddHistograms --add-nicks "mt_noplot et_noplot" --add-result-nicks sumThese three commands are equivalent, although the automatically chosen binning can make them look different. This can be changed specifying --x-bins.
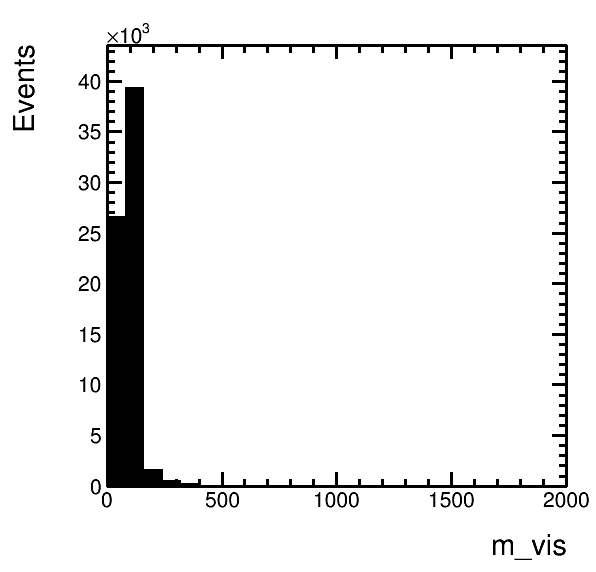

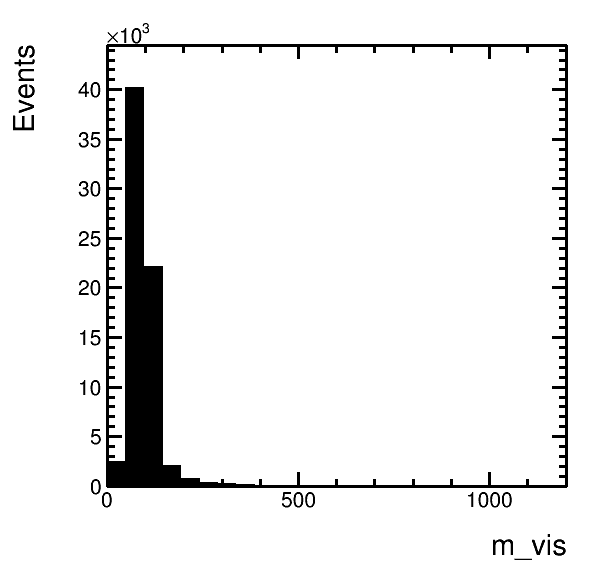
higgsplot.py -i output.root -f mt/ntuple -x eta_1 -y pt_1 --tree-draw-options prof -m E --x-label "#eta(#mu)" --y-label "p_{T}(#mu) / GeV" --title Title
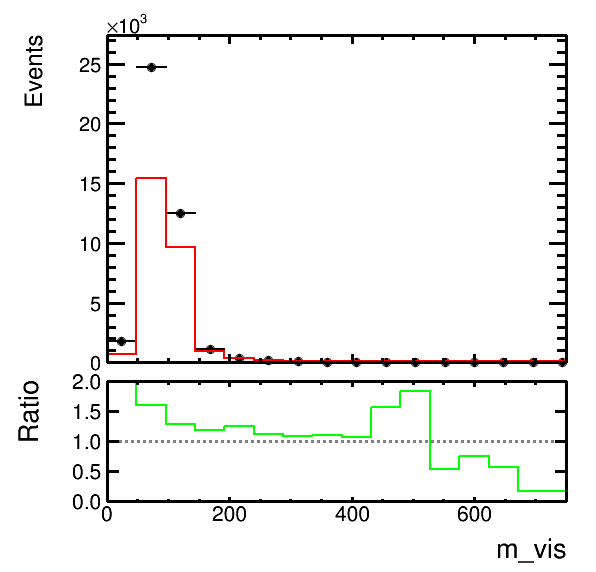
higgsplot.py -i output.root -f mt/ntuple et/ntuple -x m_vis --analysis-modules Ratio --x-label "m_{vis} / GeV" --y-subplot-label "Data/MC"higgsplot.py -i output.root -f mt/ntuple et/ntuple mt/ntuple -x m_vis -w weight weight "1.5*weight" --stacks mc mc data -m HIST HIST E E --colors kGreen kRed kBlack kBlack --nicks mc1 mc2 data --analysis-modules Ratio --ratio-numerator-nicks data --ratio-denominator-nicks "mc1 mc2" --labels "Bkg. 1" "Bkg. 2" "Data" "" --legend 0.7 0.65 0.9 0.85 --legend-markers F F ELP ""
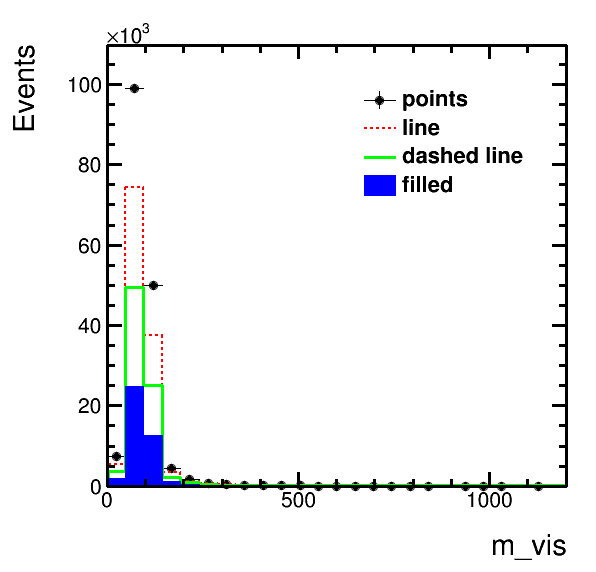
higgsplot.py -i output.root -f mt/ntuple -x m_vis -w 4 3 2 1 -m E LINE LINE HIST --line-styles 1 2 1 1 --line-widths 1 2 3 1 --legend 0.6 0.65 0.9 0.85 --legend-markers ELP L L F --labels points "dashed line" line filledhiggsplot.py -i output.root -f mt/ntuple et/ntuple mt/ntuple -x m_vis -w weight weight "1.5*weight" --stacks mc mc data -m HIST HIST E --colors "#220088" kYellow 1higgsplot.py -i output.root -f mt/ntuple -x eta_1 -y pt_1 --tree-draw-options prof -m E --y-lims 40 50
higgsplot.py -i output.root -f mt/ntuple et/ntuple -x m_vis --analysis-modules Ratio --y-subplot-lims 0 2 --x-lims 0 750