| title | author | date |
|---|---|---|
Automating Data-analysis Pipelines |
Shaun Jackman |
2014-11-03 |
| UBC STAT 545A/STAT 547M
| 2014-11-03
| Shaun Jackman @sjackman
| 
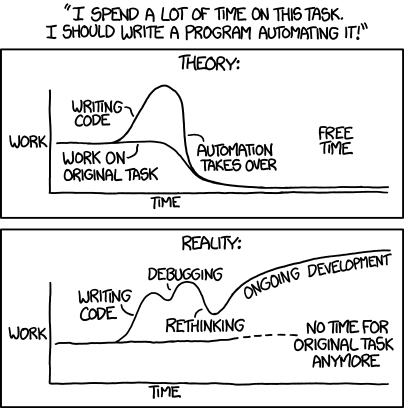
'Automating' comes from the roots 'auto-' meaning 'self-', and 'mating', meaning 'screwing'.
breaks up a monolithic make-all-the-things script into discrete, manageable chunks.
| … defines its input and its outputs. | … does not modify its inputs, so it is idempotent.
| Rerunning a stage of the pipeline | produces the same results as the previous run.
| When you modify one stage of the pipeline, | you don't have to rerun the entire pipeline.
You only rerun the downstream, dependent stages.
Divide up work amongst a group by assigning to each person stages of the pipeline design.
| You can draw pretty pictures of your pipeline, | because a pipeline is a graph.
|  | 01_justR
| 01_justR
| … to reproduce previous results. | … to recreate results deleted by fat fingers. | … to rerun the pipeline with updated software. | … to run the same pipeline on a new data set.
#!/usr/bin/env Rscript
source("00_downloadData.R")
source("01_filterReorder.R")
source("02_aggregatePlot.R"). . .
- Shows in what order to run the scripts.
- You can resume the pipeline from the middle.
#!/bin/sh
set -eux
Rscript 00_downloadData.R
Rscript 01_filterReorder.R
Rscript 02_aggregatePlot.R. . .
Allows you to easily run your pipeline from the shell.
. . .
| Option | Effect |
|---|---|
set -e |
Stop at the first error |
set -u |
Undefined variables are an error |
set -x |
Print each command as it is run |
#!/bin/sh
set -eux
curl -L http://bit.ly/lotr_raw-tsv >lotr_raw.tsv
Rscript 01_filterReorder.R
Rscript 02_aggregatePlot.R. . .
R is a good tool, but not always the best tool for the job.
Not sacrilege, but the principal tenet of a polyglot.
#!/usr/bin/make -f
lotr_raw.tsv:
curl -L http://bit.ly/lotr_raw-tsv >lotr_raw.tsv
lotr_clean.tsv: 01_filterReorder.R lotr_raw.tsv
Rscript 01_filterReorder.R
totalWordsByFilmRace.tsv: 02_aggregatePlot.R lotr_clean.tsv
Rscript 02_aggregatePlot.R. . .
| A Makefile gives both the commands | and their dependencies.
| Tell Make how to create one type of file from another | and which files you want to create.
. . .
| Make looks at which files you have | and figures out how to create the files that you want.
Scripts and data files are vertices of the graph.
Dependencies between stages are edges of the graph.
Both scripts and data files are shown.
|  | 01_justR
| 01_justR
- Only dependencies between scripts are shown.
- Data files are not shown.
- Run the scripts in topographical order.
|  | STAT 540 Differential Methylation in Leukemia
| STAT 540 Differential Methylation in Leukemia
A shell script gives one order in which you can successfully run the pipeline.
. . .
Unless the pipeline is completely linear, there are likely other such orders.
A different order of commands may be more convenient, but without information of the dependencies, you're stuck with the given order.
- Downloads the data
- Runs the command-line programs
- Performs the statistical analyses using R
- and Generates the TSV tables
- Renders the figures using ggplot2
- Renders the supplementary material using RMarkdown
- Renders the manuscript using Pandoc
Plain Text, Papers, Pandoc by Kieran Healy
Markdown is a plain-text typesetting language
A header
========
A list:
+ This text is *italic*
+ This text is **bold**A list:
- This text is italic
- This text is bold
- RMarkdown interleaves prose with R code
- to aggregate and summarize the data
- to generate tables
- to render figures using ggplot2
- RMarkdown is ideal for supplementary material
The Sum of 1 + 1
================
The sum of 1 + 1 is calculated as follows.
```{r}
1 + 1
```

The sum of 1 + 1 is calculated as follows.
1 + 1## [1] 2
Dependencies of article/Makefile
%.md: %.Rmd
Rscript -e 'knitr::knit("$<", "$@")'
%.html: %.md
pandoc -s -o $@ $<
%.html: %.Rmd
Rscript -e 'rmarkdown::render("$<")'
article.html: figure.png
%.png: %.gv
dot -Tpng $< >$@make article.htmldot -Tpng figure.gv >figure.png
Rscript -e 'rmarkdown::render("article.Rmd")'%.md: %.Rmd
Rscript -e 'knitr::knit("$<", "$@")'
%.html: %.md
pandoc -s -o $@ $<
%.html: %.Rmd
Rscript -e 'rmarkdown::render("$<")'
article.html: figure.png
%.png: %.gv
dot -Tpng $< >$@make article.md article.htmlRscript -e 'knitr::knit("article.Rmd", "article.md")'
dot -Tpng figure.gv >figure.png
pandoc -s -o article.html article.md| Pandoc renders attractive documents and slides | from plain-text typesetting formats
It converts between every format known (just about)
- Markdown
- HTML
- LaTeX
- ODT and docx (yes, really)
#!/bin/sh
set -eux
dot -Tpng -o figure.png figure.gv
Rscript -e 'knitr::knit("article.Rmd")'
pandoc -s -o article.html article.mdShell script
all:
dot -Tpng -o figure.png figure.gv
Rscript -e 'knitr::knit("article.Rmd")'
pandoc -s -o article.html article.mdFirst Makefile
all: article.html
article.html:
dot -Tpng -o figure.png figure.gv
Rscript -e 'knitr::knit("article.Rmd")'
pandoc -s -o article.html article.mdAdd a rule to build article.html
all: article.html
article.html: article.Rmd
dot -Tpng -o figure.png figure.gv
Rscript -e 'knitr::knit("article.Rmd")'
pandoc -s -o article.html article.mdarticle.html depends on article.Rmd
all: article.html
figure.png: figure.gv
dot -Tpng -o figure.png figure.gv
article.md: article.Rmd
Rscript -e 'knitr::knit("article.Rmd")'
article.html: article.md figure.png
pandoc -s -o article.html article.mdSplit one rule into three
all: article.html
figure.png: figure.gv
dot -Tpng -o $@ $<
article.md: article.Rmd
Rscript -e 'knitr::knit("$<", "$@")'
article.html: article.md figure.png
pandoc -s -o $@ $<Use the variables $< and $@ for the input and output file
all: article.html
%.png: %.gv
dot -Tpng -o $@ $<
%.md: %.Rmd
Rscript -e 'knitr::knit("$<", "$@")'
article.html: article.md figure.png
pandoc -s -o $@ $<Use pattern rules. The % matches any string
all: article.html
%.png: %.gv
dot -Tpng -o $@ $<
%.md: %.Rmd
Rscript -e 'knitr::knit("$<", "$@")'
%.html: %.md
pandoc -s -o $@ $<
article.html: figure.pngarticle.html also depends on figure.png
all: article.html
clean:
rm -f article.md article.html figure.png
%.png: %.gv
dot -Tpng -o $@ $<
%.md: %.Rmd
Rscript -e 'knitr::knit("$<", "$@")'
%.html: %.md
pandoc -s -o $@ $<
article.html: figure.pngAdd the target named clean
all: article.html
clean:
rm -f article.md article.html figure.png
.PHONY: all clean
.DELETE_ON_ERROR:
.SECONDARY:
%.png: %.gv
dot -Tpng -o $@ $<
%.md: %.Rmd
Rscript -e 'knitr::knit("$<", "$@")'
%.html: %.md
pandoc -s -o $@ $<
article.html: figure.pngAdd .PHONY, .DELETE_ON_ERROR and .SECONDARY
all: article.html
clean:
rm -f article.md article.html figure.png
.PHONY: all clean
.DELETE_ON_ERROR:
.SECONDARY:
# Render a GraphViz file
%.png: %.gv
dot -Tpng -o $@ $<
# Knit a RMarkdown document
%.md: %.Rmd
Rscript -e 'knitr::knit("$<", "$@")'
# Render a Markdown document to HTML
%.html: %.md
pandoc -s -o $@ $<
# Dependencies on figures
article.html: figure.png| STAT 545A | xkcd automation | R | Rscript | shell | make | Markdown | RMarkdown | Pandoc | ggplot2 | Plain Text, Papers, Pandoc | STAT 540 Differential Methylation in Leukemia
@sjackman | github.com/sjackman | sjackman.ca
| Genome Sciences Centre, BC Cancer Agency | Vancouver, Canada | @sjackman | github.com/sjackman | sjackman.ca