diff --git a/README.md b/README.md
index a2ca129e5..3766e918a 100644
--- a/README.md
+++ b/README.md
@@ -1,15 +1,15 @@
-[](https://github.com/pre-commit/pre-commit) [](https://github.com/ilabcode/pyhgf/blob/master/LICENSE) [](https://codecov.io/gh/ilabcode/pyhgf) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/) [](https://badge.fury.io/py/pyhgf)
+[](https://github.com/pre-commit/pre-commit) [](https://github.com/ComputationalPsychiatry/pyhgf/blob/master/LICENSE) [](https://codecov.io/gh/ComputationalPsychiatry/pyhgf) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/) [](https://badge.fury.io/py/pyhgf)
# PyHGF: A Neural Network Library for Predictive Coding
- +
+ PyHGF is a Python library for creating and manipulating dynamic probabilistic networks for predictive coding. These networks approximate Bayesian inference by optimizing beliefs through the diffusion of predictions and precision-weighted prediction errors. The network structure remains flexible during message-passing steps, allowing for dynamic adjustments. They can be used as a biologically plausible cognitive model in computational neuroscience or as a generalization of Bayesian filtering for designing efficient, modular decision-making agents. The default implementation supports the generalized Hierarchical Gaussian Filters (gHGF, Weber et al., 2024), but the framework is designed to be adaptable to other algorithms. Built on top of JAX, the core functions are differentiable and JIT-compiled where applicable. The library is optimized for modularity and ease of use, allowing seamless integration with other libraries in the ecosystem for Bayesian inference and optimization. Additionally, a binding with an implementation in Rust is under active development, which will further enhance flexibility during inference. You can find the method paper describing the toolbox [here](https://arxiv.org/abs/2410.09206) and the method paper describing the gHGF, which is the main framework currently supported by the toolbox [here](https://arxiv.org/abs/2305.10937).
-* 📖 [API Documentation](https://ilabcode.github.io/pyhgf/api.html)
-* ✏️ [Tutorials and examples](https://ilabcode.github.io/pyhgf/learn.html)
+* 📖 [API Documentation](https://computationalpsychiatry.github.io/pyhgf/api.html)
+* ✏️ [Tutorials and examples](https://computationalpsychiatryionalpsychiatry.github.io/pyhgf/learn.html)
## Getting started
@@ -21,7 +21,7 @@ The last official release can be downloaded from PIP:
The current version under development can be installed from the master branch of the GitHub folder:
-`pip install “git+https://github.com/ilabcode/pyhgf.git”`
+`pip install “git+https://github.com/ComputationalPsychiatry/pyhgf.git”`
### How does it work?
@@ -32,12 +32,12 @@ Dynamic networks can be defined as a tuple containing the following variables:
* A set of update functions. An update function receive a network tuple and returns an updated network tuple.
* An update sequence (tuple) that defines the order and target of the update functions.
-
PyHGF is a Python library for creating and manipulating dynamic probabilistic networks for predictive coding. These networks approximate Bayesian inference by optimizing beliefs through the diffusion of predictions and precision-weighted prediction errors. The network structure remains flexible during message-passing steps, allowing for dynamic adjustments. They can be used as a biologically plausible cognitive model in computational neuroscience or as a generalization of Bayesian filtering for designing efficient, modular decision-making agents. The default implementation supports the generalized Hierarchical Gaussian Filters (gHGF, Weber et al., 2024), but the framework is designed to be adaptable to other algorithms. Built on top of JAX, the core functions are differentiable and JIT-compiled where applicable. The library is optimized for modularity and ease of use, allowing seamless integration with other libraries in the ecosystem for Bayesian inference and optimization. Additionally, a binding with an implementation in Rust is under active development, which will further enhance flexibility during inference. You can find the method paper describing the toolbox [here](https://arxiv.org/abs/2410.09206) and the method paper describing the gHGF, which is the main framework currently supported by the toolbox [here](https://arxiv.org/abs/2305.10937).
-* 📖 [API Documentation](https://ilabcode.github.io/pyhgf/api.html)
-* ✏️ [Tutorials and examples](https://ilabcode.github.io/pyhgf/learn.html)
+* 📖 [API Documentation](https://computationalpsychiatry.github.io/pyhgf/api.html)
+* ✏️ [Tutorials and examples](https://computationalpsychiatryionalpsychiatry.github.io/pyhgf/learn.html)
## Getting started
@@ -21,7 +21,7 @@ The last official release can be downloaded from PIP:
The current version under development can be installed from the master branch of the GitHub folder:
-`pip install “git+https://github.com/ilabcode/pyhgf.git”`
+`pip install “git+https://github.com/ComputationalPsychiatry/pyhgf.git”`
### How does it work?
@@ -32,12 +32,12 @@ Dynamic networks can be defined as a tuple containing the following variables:
* A set of update functions. An update function receive a network tuple and returns an updated network tuple.
* An update sequence (tuple) that defines the order and target of the update functions.
- +
+ You can find a deeper introduction to how to create and manipulate networks under the following link:
-* 🎓 [Creating and manipulating networks of probabilistic nodes](https://ilabcode.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
+* 🎓 [Creating and manipulating networks of probabilistic nodes](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
### The Generalized Hierarchical Gaussian Filter
@@ -46,7 +46,7 @@ Generalized Hierarchical Gaussian Filters (gHGF) are specific instances of dynam
You can find a deeper introduction on how does the gHGF works under the following link:
-* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
+* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
### Model fitting
@@ -73,7 +73,7 @@ hgf.input_data(input_data=u)
hgf.plot_trajectories();
```
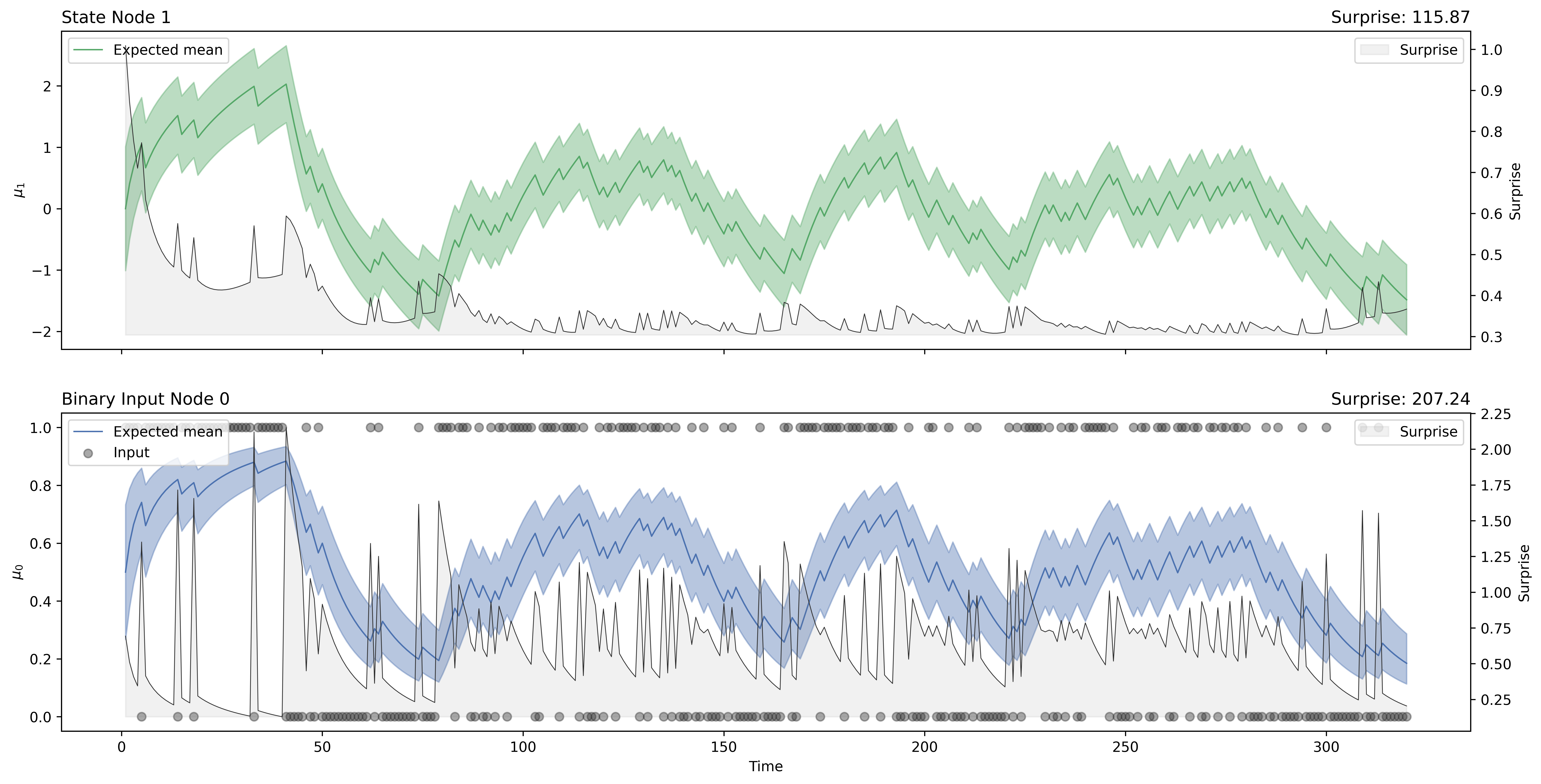
-
+
```python
from pyhgf.response import binary_softmax_inverse_temperature
@@ -93,7 +93,7 @@ print(f"Sum of surprises = {surprise.sum()}")
## Acknowledgments
-This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ilabcode/HGF.jl).
+This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ComputationalPsychiatry/HGF.jl).
## References
diff --git a/docs/source/conf.py b/docs/source/conf.py
index 839ef49ab..3dd718eab 100644
--- a/docs/source/conf.py
+++ b/docs/source/conf.py
@@ -95,7 +95,7 @@
"icon_links": [
dict(
name="GitHub",
- url="https://github.com/ilabcode/pyhgf",
+ url="https://github.com/ComputationalPsychiatry/pyhgf",
icon="fa-brands fa-square-github",
),
dict(
diff --git a/docs/source/index.md b/docs/source/index.md
index 6d8b78ef7..95225a926 100644
--- a/docs/source/index.md
+++ b/docs/source/index.md
@@ -1,14 +1,14 @@
-[](https://github.com/pre-commit/pre-commit) [](https://github.com/ilabcode/pyhgf/blob/master/LICENSE) [](https://codecov.io/gh/ilabcode/pyhgf) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/) [](https://badge.fury.io/py/pyhgf)
+[](https://github.com/pre-commit/pre-commit) [](https://github.com/ComputationalPsychiatry/pyhgf/blob/master/LICENSE) [](https://codecov.io/gh/ComputationalPsychiatry/pyhgf) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/) [](https://badge.fury.io/py/pyhgf)
# PyHGF: A Neural Network Library for Predictive Coding
-
You can find a deeper introduction to how to create and manipulate networks under the following link:
-* 🎓 [Creating and manipulating networks of probabilistic nodes](https://ilabcode.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
+* 🎓 [Creating and manipulating networks of probabilistic nodes](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
### The Generalized Hierarchical Gaussian Filter
@@ -46,7 +46,7 @@ Generalized Hierarchical Gaussian Filters (gHGF) are specific instances of dynam
You can find a deeper introduction on how does the gHGF works under the following link:
-* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
+* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
### Model fitting
@@ -73,7 +73,7 @@ hgf.input_data(input_data=u)
hgf.plot_trajectories();
```
-
+
```python
from pyhgf.response import binary_softmax_inverse_temperature
@@ -93,7 +93,7 @@ print(f"Sum of surprises = {surprise.sum()}")
## Acknowledgments
-This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ilabcode/HGF.jl).
+This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ComputationalPsychiatry/HGF.jl).
## References
diff --git a/docs/source/conf.py b/docs/source/conf.py
index 839ef49ab..3dd718eab 100644
--- a/docs/source/conf.py
+++ b/docs/source/conf.py
@@ -95,7 +95,7 @@
"icon_links": [
dict(
name="GitHub",
- url="https://github.com/ilabcode/pyhgf",
+ url="https://github.com/ComputationalPsychiatry/pyhgf",
icon="fa-brands fa-square-github",
),
dict(
diff --git a/docs/source/index.md b/docs/source/index.md
index 6d8b78ef7..95225a926 100644
--- a/docs/source/index.md
+++ b/docs/source/index.md
@@ -1,14 +1,14 @@
-[](https://github.com/pre-commit/pre-commit) [](https://github.com/ilabcode/pyhgf/blob/master/LICENSE) [](https://codecov.io/gh/ilabcode/pyhgf) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/) [](https://badge.fury.io/py/pyhgf)
+[](https://github.com/pre-commit/pre-commit) [](https://github.com/ComputationalPsychiatry/pyhgf/blob/master/LICENSE) [](https://codecov.io/gh/ComputationalPsychiatry/pyhgf) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/) [](https://badge.fury.io/py/pyhgf)
# PyHGF: A Neural Network Library for Predictive Coding
- +
+ PyHGF is a Python library for creating and manipulating dynamic probabilistic networks for predictive coding. These networks approximate Bayesian inference by optimizing beliefs through the diffusion of predictions and precision-weighted prediction errors. The network structure remains flexible during message-passing steps, allowing for dynamic adjustments. They can be used as a biologically plausible cognitive model in computational neuroscience or as a generalization of Bayesian filtering for designing efficient, modular decision-making agents. The default implementation supports the generalized Hierarchical Gaussian Filters (gHGF, Weber et al., 2024), but the framework is designed to be adaptable to other algorithms. Built on top of JAX, the core functions are differentiable and JIT-compiled where applicable. The library is optimized for modularity and ease of use, allowing seamless integration with other libraries in the ecosystem for Bayesian inference and optimization. Additionally, a binding with an implementation in Rust is under active development, which will further enhance flexibility during inference. You can find the method paper describing the toolbox [here](https://arxiv.org/abs/2410.09206) and the method paper describing the gHGF, which is the main framework currently supported by the toolbox [here](https://arxiv.org/abs/2305.10937).
-* 📖 [API Documentation](https://ilabcode.github.io/pyhgf/api.html)
-* ✏️ [Tutorials and examples](https://ilabcode.github.io/pyhgf/learn.html)
+* 📖 [API Documentation](https://computationalpsychiatry.github.io/pyhgf/api.html)
+* ✏️ [Tutorials and examples](https://computationalpsychiatry.github.io/pyhgf/learn.html)
## Getting started
@@ -23,7 +23,7 @@ pip install pyhgf
The current version under development can be installed from the master branch of the GitHub folder:
```bash
-pip install “git+https://github.com/ilabcode/pyhgf.git”
+pip install “git+https://github.com/ComputationalPsychiatry/pyhgf.git”
```
### How does it work?
@@ -35,12 +35,12 @@ Dynamic networks can be defined as a tuple containing the following variables:
* A set of update functions. An update function receive a network tuple and returns an updated network tuple.
* An update sequence (tuple) that defines the order and target of the update functions.
-
PyHGF is a Python library for creating and manipulating dynamic probabilistic networks for predictive coding. These networks approximate Bayesian inference by optimizing beliefs through the diffusion of predictions and precision-weighted prediction errors. The network structure remains flexible during message-passing steps, allowing for dynamic adjustments. They can be used as a biologically plausible cognitive model in computational neuroscience or as a generalization of Bayesian filtering for designing efficient, modular decision-making agents. The default implementation supports the generalized Hierarchical Gaussian Filters (gHGF, Weber et al., 2024), but the framework is designed to be adaptable to other algorithms. Built on top of JAX, the core functions are differentiable and JIT-compiled where applicable. The library is optimized for modularity and ease of use, allowing seamless integration with other libraries in the ecosystem for Bayesian inference and optimization. Additionally, a binding with an implementation in Rust is under active development, which will further enhance flexibility during inference. You can find the method paper describing the toolbox [here](https://arxiv.org/abs/2410.09206) and the method paper describing the gHGF, which is the main framework currently supported by the toolbox [here](https://arxiv.org/abs/2305.10937).
-* 📖 [API Documentation](https://ilabcode.github.io/pyhgf/api.html)
-* ✏️ [Tutorials and examples](https://ilabcode.github.io/pyhgf/learn.html)
+* 📖 [API Documentation](https://computationalpsychiatry.github.io/pyhgf/api.html)
+* ✏️ [Tutorials and examples](https://computationalpsychiatry.github.io/pyhgf/learn.html)
## Getting started
@@ -23,7 +23,7 @@ pip install pyhgf
The current version under development can be installed from the master branch of the GitHub folder:
```bash
-pip install “git+https://github.com/ilabcode/pyhgf.git”
+pip install “git+https://github.com/ComputationalPsychiatry/pyhgf.git”
```
### How does it work?
@@ -35,12 +35,12 @@ Dynamic networks can be defined as a tuple containing the following variables:
* A set of update functions. An update function receive a network tuple and returns an updated network tuple.
* An update sequence (tuple) that defines the order and target of the update functions.
- +
+ You can find a deeper introduction to how to create and manipulate networks under the following link:
-* 🎓 [Creating and manipulating networks of probabilistic nodes](https://ilabcode.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
+* 🎓 [Creating and manipulating networks of probabilistic nodes](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
### The Generalized Hierarchical Gaussian Filter
@@ -49,7 +49,7 @@ Generalized Hierarchical Gaussian Filters (gHGF) are specific instances of dynam
You can find a deeper introduction on how does the gHGF works under the following link:
-* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
+* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
### Model fitting
@@ -76,7 +76,7 @@ hgf.input_data(input_data=u)
hgf.plot_trajectories();
```
-
+
```python
from pyhgf.response import binary_softmax_inverse_temperature
@@ -96,7 +96,7 @@ print(f"Sum of surprises = {surprise.sum()}")
## Acknowledgments
-This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ilabcode/HGF.jl).
+This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ComputationalPsychiatry/HGF.jl).
## References
diff --git a/docs/source/learn.md b/docs/source/learn.md
index f99e8672c..076e1f8cb 100644
--- a/docs/source/learn.md
+++ b/docs/source/learn.md
@@ -50,7 +50,7 @@ notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
# Learn
-In this section, you can find tutorial notebooks that describe the internals of pyhgf, the theory behind the Hierarchical Gaussian filter, and step-by-step application and use cases of the model. At the beginning of every tutorial, you will find a badge [](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.1-Creating_networks.ipynb) to run the notebook interactively in a Google Colab session.
+In this section, you can find tutorial notebooks that describe the internals of pyhgf, the theory behind the Hierarchical Gaussian filter, and step-by-step application and use cases of the model. At the beginning of every tutorial, you will find a badge [](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.1-Creating_networks.ipynb) to run the notebook interactively in a Google Colab session.
## Theory
diff --git a/docs/source/notebooks/0.1-Theory.ipynb b/docs/source/notebooks/0.1-Theory.ipynb
index c995adf49..af2327fbe 100644
--- a/docs/source/notebooks/0.1-Theory.ipynb
+++ b/docs/source/notebooks/0.1-Theory.ipynb
@@ -14,7 +14,7 @@
"(theory)=\n",
"# Introduction to the Generalised Hierarchical Gaussian Filter\n",
"\n",
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.1-Theory.ipynb)\n",
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.1-Theory.ipynb)\n",
"\n",
"The Hierarchical Gaussian Filter is a Bayesian model of belief updating under uncertainty in which the volatility of the environment is encoded by a hierarchy of probability distributions. A generalised version of this model {cite:p}`weber:2023` has further framed this into a neural network framework of probabilistic state nodes interacting with each other. At the heart of both frameworks lies a generative model that consist in a hierarchy of {term}`Gaussian Random Walk`. The inversion of this model (i.e. estimating parameters from observed values) leads to the derivation of prediction errors and posterior updates that can turn these networks into learning systems. To fully understand this model, it is therefore central to start with the simplest case, the implied generative model, which can be seen as the probabilistic structure that generates observations.\n",
"\n",
@@ -664,7 +664,7 @@
"source": [
"### The propagation of prediction and prediction errors\n",
"\n",
- "Having described the model as a specific configuration of predictive nodes offer many advantages, especially in term of modularity for the user. However, the model itself is not limited to the description of the generative process that we covered in the previous examples. The most interesting, and also the more complex, part of the modelling consists of the capability for the network to update the hierarchical structure in a Bayesian optimal way as new observations are presented. These steps are defined by a set of simple, one-step update equations that represent changes in beliefs about the hidden states (i.e. the sufficient statistics of the nodes) specified in the generative model. These equations were first described in {cite:t}`2011:mathys`, and the update equations for volatility and value coupling in the generalized Hierarchical Gaussian filter (on which most of the update functions in [pyhgf](https://github.com/ilabcode/pyhgf) are based) have been described in {cite:p}`weber:2023`. The exact computations in each step especially depend on the nature of the coupling (via {term}`VAPE`s vs. {term}`VOPE`s) between the parent and children nodes. It is beyond the scope of this tutorial to dive into the derivation of these steps and we refer the interested reader to the mentioned papers. Here, we provide a general overview of the dynamic of the update sequence that supports belief updating. The computations triggered by any observation at each time point can be ordered in time as shown in the belief update algorithm."
+ "Having described the model as a specific configuration of predictive nodes offer many advantages, especially in term of modularity for the user. However, the model itself is not limited to the description of the generative process that we covered in the previous examples. The most interesting, and also the more complex, part of the modelling consists of the capability for the network to update the hierarchical structure in a Bayesian optimal way as new observations are presented. These steps are defined by a set of simple, one-step update equations that represent changes in beliefs about the hidden states (i.e. the sufficient statistics of the nodes) specified in the generative model. These equations were first described in {cite:t}`2011:mathys`, and the update equations for volatility and value coupling in the generalized Hierarchical Gaussian filter (on which most of the update functions in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) are based) have been described in {cite:p}`weber:2023`. The exact computations in each step especially depend on the nature of the coupling (via {term}`VAPE`s vs. {term}`VOPE`s) between the parent and children nodes. It is beyond the scope of this tutorial to dive into the derivation of these steps and we refer the interested reader to the mentioned papers. Here, we provide a general overview of the dynamic of the update sequence that supports belief updating. The computations triggered by any observation at each time point can be ordered in time as shown in the belief update algorithm."
]
},
{
diff --git a/docs/source/notebooks/0.2-Creating_networks.ipynb b/docs/source/notebooks/0.2-Creating_networks.ipynb
index 82dffc43c..c3640e0e5 100644
--- a/docs/source/notebooks/0.2-Creating_networks.ipynb
+++ b/docs/source/notebooks/0.2-Creating_networks.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.2-Creating_networks.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.2-Creating_networks.ipynb)"
]
},
{
@@ -85,7 +85,7 @@
"id": "57dde3f9-d8b1-437e-8a33-7aea1a1b1e2e",
"metadata": {},
"source": [
- "[pyhgf](https://ilabcode.github.io/pyhgf/index.html#) is designed with inspiration from graph neural network libraries that can support message-passing schemes and perform belief propagation through networks of probabilistic nodes. Here, this principle is applied to predictive processing and focuses on networks that are structured as **rooted trees** and perform variational message passing to update beliefs about the state of the environment, inferred from the observations at the root of the tree. While this library is optimized to implement the standard two-level and three-level HGF {cite:p}`2011:mathys,2014:mathys`, as well as the generalized HGF {cite:p}`weber:2023`, it can also be applied to much larger use cases, with the idea is to generalize belief propagation as it has been described so far to larger and more complex networks that will capture a greater variety of environmental structure. Therefore, the library is also designed to facilitate the creation and manipulation of such probabilistic networks. Importantly, here we consider that a probabilistic network should be defined by the following four variables:\n",
+ "[pyhgf](https://computationalpsychiatry.github.io/pyhgf/index.html#) is designed with inspiration from graph neural network libraries that can support message-passing schemes and perform belief propagation through networks of probabilistic nodes. Here, this principle is applied to predictive processing and focuses on networks that are structured as **rooted trees** and perform variational message passing to update beliefs about the state of the environment, inferred from the observations at the root of the tree. While this library is optimized to implement the standard two-level and three-level HGF {cite:p}`2011:mathys,2014:mathys`, as well as the generalized HGF {cite:p}`weber:2023`, it can also be applied to much larger use cases, with the idea is to generalize belief propagation as it has been described so far to larger and more complex networks that will capture a greater variety of environmental structure. Therefore, the library is also designed to facilitate the creation and manipulation of such probabilistic networks. Importantly, here we consider that a probabilistic network should be defined by the following four variables:\n",
"1. the network parameters\n",
"2. the network structure\n",
"3. the update function(s)\n",
@@ -711,7 +711,7 @@
"source": [
"### Multivariate coupling\n",
"\n",
- "As we can see in the examples above, nodes in a valid HGF network can be influenced by multiple parents (either value or volatility parents). Similarly, a single node can be influenced by multiple children. This feature is termed *multivariate descendency* and *multivariate ascendency* (respectively) and is a central addition to the generalization of the HGF {cite:p}`weber:2023` that was implemented in this package, as well as in the [Julia counterpart](https://github.com/ilabcode/HierarchicalGaussianFiltering.jl)."
+ "As we can see in the examples above, nodes in a valid HGF network can be influenced by multiple parents (either value or volatility parents). Similarly, a single node can be influenced by multiple children. This feature is termed *multivariate descendency* and *multivariate ascendency* (respectively) and is a central addition to the generalization of the HGF {cite:p}`weber:2023` that was implemented in this package, as well as in the [Julia counterpart](https://github.com/ComputationalPsychiatry/HierarchicalGaussianFiltering.jl)."
]
},
{
@@ -1748,7 +1748,7 @@
"source": [
"## Creating custom update functions\n",
"\n",
- "The structure of the network and the node's parameters are the most static component of the network. Actually, we could consider that the network already exists once those two variables are in place. However in [pyhgf](https://ilabcode.github.io/pyhgf/index.html#) we consider that the update functions $\\mathcal{F} = \\{f_1, ..., f_n\\}$ and the update sequence $\\Sigma = [f_1(n_1), ..., f_i(n_j), f \\in \\mathcal{F}, n \\in 1, ..., k ]$ (the order of update) are also part of the models. This choice was made to explicitly account that there is no one unique way of modelling the way beliefs propagate through the network, and a core task for predictive coding applications is to develop new *probabilistic nodes* that account for a greater variety of phenomena. This step critically requires modelling beliefs diffusion, and therefore to modify, or creating the underlying update functions."
+ "The structure of the network and the node's parameters are the most static component of the network. Actually, we could consider that the network already exists once those two variables are in place. However in [pyhgf](https://computationalpsychiatry.github.io/pyhgf/index.html#) we consider that the update functions $\\mathcal{F} = \\{f_1, ..., f_n\\}$ and the update sequence $\\Sigma = [f_1(n_1), ..., f_i(n_j), f \\in \\mathcal{F}, n \\in 1, ..., k ]$ (the order of update) are also part of the models. This choice was made to explicitly account that there is no one unique way of modelling the way beliefs propagate through the network, and a core task for predictive coding applications is to develop new *probabilistic nodes* that account for a greater variety of phenomena. This step critically requires modelling beliefs diffusion, and therefore to modify, or creating the underlying update functions."
]
},
{
diff --git a/docs/source/notebooks/0.3-Generalised_filtering.ipynb b/docs/source/notebooks/0.3-Generalised_filtering.ipynb
index 552989459..5452eb9ce 100644
--- a/docs/source/notebooks/0.3-Generalised_filtering.ipynb
+++ b/docs/source/notebooks/0.3-Generalised_filtering.ipynb
@@ -14,7 +14,7 @@
"(generalised_filtering)=\n",
"# From Reinforcement Learning to Generalised Bayesian Filtering\n",
"\n",
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.3-Generalised_filtering.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.3-Generalised_filtering.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/1.1-Binary_HGF.ipynb b/docs/source/notebooks/1.1-Binary_HGF.ipynb
index 83297bccc..5d02d4deb 100644
--- a/docs/source/notebooks/1.1-Binary_HGF.ipynb
+++ b/docs/source/notebooks/1.1-Binary_HGF.ipynb
@@ -20,7 +20,7 @@
"id": "1258ea53-11fc-4a27-927c-2afc91f4aa12",
"metadata": {},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.1-Binary_HGF.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.1-Binary_HGF.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/1.2-Categorical_HGF.ipynb b/docs/source/notebooks/1.2-Categorical_HGF.ipynb
index f4a42850f..06ab288d2 100644
--- a/docs/source/notebooks/1.2-Categorical_HGF.ipynb
+++ b/docs/source/notebooks/1.2-Categorical_HGF.ipynb
@@ -20,7 +20,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.3-CAtegorical_HGF.ipynb)\n",
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.3-CAtegorical_HGF.ipynb)\n",
"\n",
"```{warning}\n",
"The categorical state node and the categorical state-transition nodes are still work in progress. The examples provided here are given for illustration. Things may change or not work until the official publication.\n",
diff --git a/docs/source/notebooks/1.3-Continuous_HGF.ipynb b/docs/source/notebooks/1.3-Continuous_HGF.ipynb
index 84e0def7e..db1e94681 100644
--- a/docs/source/notebooks/1.3-Continuous_HGF.ipynb
+++ b/docs/source/notebooks/1.3-Continuous_HGF.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.3-Continuous_HGF.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.3-Continuous_HGF.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/2-Using_custom_response_functions.ipynb b/docs/source/notebooks/2-Using_custom_response_functions.ipynb
index 71554bcb4..774e43dad 100644
--- a/docs/source/notebooks/2-Using_custom_response_functions.ipynb
+++ b/docs/source/notebooks/2-Using_custom_response_functions.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/2-Using_custom_response_functions.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/2-Using_custom_response_functions.ipynb)"
]
},
{
@@ -542,12 +542,12 @@
"\n",
"Let's now consider that the two vectors of observations $u$ and responses $y$ were obtained from a real participant undergoing a real experiment. In this situation, we assume that this participant internally used a {term}`Perceptual model` and a {term}`Decision rule` that might resemble what we defined previously, and we want to infer what are the most likely values for critical parameters in the model (e.g. the evolution rate $\\omega_2$). To do so, we are going to use our dataset (both $u$ and $y$), and try many models. We are going to fix the values of all HGF parameters to reasonable estimates (here, using the exact same values as in the simulation), except for $\\omega_2$. For this last parameter, we will assume a prior set at $\\mathcal{N}(-2.0, 2.0)$. The idea is that we want to sample many $\\omega_2$ values from this distribution and see if the model is performing better with some values.\n",
"\n",
- "But here, we need a clear definition of what this means *to perform better* for a given model. And this is exactly what a {term}`Response model` does, it is a way for us to evaluate how likely the behaviours $y$ for a given {term}`Perceptual model`, and assuming that the participants use this specific {term}`Decision rule`. In [pyhgf](https://github.com/ilabcode/pyhgf), this step is performed by creating the corresponding {term}`Response function`, which is the Python function that will return the surprise $S$ of getting these behaviours from the participant under this decision rule.\n",
+ "But here, we need a clear definition of what this means *to perform better* for a given model. And this is exactly what a {term}`Response model` does, it is a way for us to evaluate how likely the behaviours $y$ for a given {term}`Perceptual model`, and assuming that the participants use this specific {term}`Decision rule`. In [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), this step is performed by creating the corresponding {term}`Response function`, which is the Python function that will return the surprise $S$ of getting these behaviours from the participant under this decision rule.\n",
"\n",
"````{hint} What is a *response function*?\n",
"Most of the work around HGFs consists in creating and adapting {term}`Response model` to work with a given experimental design. There is no limit in terms of what can be used as a {term}`Response model`, provided that the {term}`Perceptual model` and the {term}`Decision rule` are clearly defined.\n",
"\n",
- "In [pyhgf](https://github.com/ilabcode/pyhgf), the {term}`Perceptual model` is the probabilistic network created with the main {py:class}`pyhgf.model.HGF` and {py:class}`pyhgf.distribution.HGFDistribution` classes. The {term}`Response model` is something that is implicitly defined when we create the {term}`Response function`, a Python function that computes the negative of the log-likelihood of the actions given the perceptual model. This {term}`Response function` can be passed as an argument to the main classes using the keywords arguments `response_function`, `response_function_inputs` and `response_function_parameters`. The `response_function` can be any callable that returns the surprise $S$ of observing action $y$ given this model, and the {term}`Decision rule`. The `response_function_inputs` are the additional data to the response function (optional) while `response_function_parameters` are the additional parameters (optional). The `response_function_inputs` is where the actions $y$ should be provided.\n",
+ "In [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), the {term}`Perceptual model` is the probabilistic network created with the main {py:class}`pyhgf.model.HGF` and {py:class}`pyhgf.distribution.HGFDistribution` classes. The {term}`Response model` is something that is implicitly defined when we create the {term}`Response function`, a Python function that computes the negative of the log-likelihood of the actions given the perceptual model. This {term}`Response function` can be passed as an argument to the main classes using the keywords arguments `response_function`, `response_function_inputs` and `response_function_parameters`. The `response_function` can be any callable that returns the surprise $S$ of observing action $y$ given this model, and the {term}`Decision rule`. The `response_function_inputs` are the additional data to the response function (optional) while `response_function_parameters` are the additional parameters (optional). The `response_function_inputs` is where the actions $y$ should be provided.\n",
"\n",
"```{important}\n",
"A *response function* should not return the actions given perceptual inputs $y | u$ (this is what the {term}`Decision rule` does), but the [surprise](https://en.wikipedia.org/wiki/Information_content) $S$ associated with the observation of actions given the perceptual inputs $S(y | u)$, which is defined by:\n",
@@ -559,7 +559,7 @@
"$$\n",
"```\n",
"\n",
- "If you are already familiar with using HGFs in the Julia equivalent of [pyhgf](https://github.com/ilabcode/pyhgf), you probably noted that the toolbox is split into a **perceptual** package [HierarchicalGaussianFiltering.jl](https://github.com/ilabcode/HierarchicalGaussianFiltering.jl) and a **response** package [ActionModels.jl](https://github.com/ilabcode/ActionModels.jl). This was made to make the difference between the two parts of the HGF clear and be explicit that you can use a perceptual model without any action model. In [pyhgf](https://github.com/ilabcode/pyhgf) however, everything happens in the same package, the response function is merely an optional, additional argument that can be passed to describe how surprise is computed.\n",
+ "If you are already familiar with using HGFs in the Julia equivalent of [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), you probably noted that the toolbox is split into a **perceptual** package [HierarchicalGaussianFiltering.jl](https://github.com/ComputationalPsychiatry/HierarchicalGaussianFiltering.jl) and a **response** package [ActionModels.jl](https://github.com/ComputationalPsychiatry/ActionModels.jl). This was made to make the difference between the two parts of the HGF clear and be explicit that you can use a perceptual model without any action model. In [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) however, everything happens in the same package, the response function is merely an optional, additional argument that can be passed to describe how surprise is computed.\n",
"````"
]
},
@@ -637,7 +637,7 @@
},
"source": [
"```{note}\n",
- "Note here that our {term}`Response function` has a structure that is the standard way to write response functions in [pyhgf](https://github.com/ilabcode/pyhgf), that is with two input arguments:\n",
+ "Note here that our {term}`Response function` has a structure that is the standard way to write response functions in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), that is with two input arguments:\n",
"- the HGF model on which the response function applies (i.e. the {term}`Perceptual model`)\n",
"- the additional parameters provided to the response function. This can include additional parameters that can be part of the equation of the model, or the input data used by this model. We then provide the `response` vector ($y$) here.\n",
"\n",
@@ -1014,7 +1014,7 @@
"```{glossary}\n",
"\n",
"Perceptual model\n",
- " The perceptual model of a Hierarchical Gaussian Filter traditionally refers to the branch receiving observations $u$ about states of the world and that performs the updating of beliefs about these states. By generalisation, the perceptual model is any probabilistic network that can be created in [pyhgf](https://github.com/ilabcode/pyhgf), receiving an arbitrary number of inputs. An HGF that only consists of a perceptual model will act as a Bayesian filter.\n",
+ " The perceptual model of a Hierarchical Gaussian Filter traditionally refers to the branch receiving observations $u$ about states of the world and that performs the updating of beliefs about these states. By generalisation, the perceptual model is any probabilistic network that can be created in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), receiving an arbitrary number of inputs. An HGF that only consists of a perceptual model will act as a Bayesian filter.\n",
"\n",
"Response model\n",
" The response model of a Hierarchical Gaussian filter refers to the branch that uses the beliefs about the state of the world to generate actions using the {term}`Decision rule`. This branch is also sometimes referred to as the **decision model** or the **observation model**, depending on the fields. Critically, this part of the model can return the surprise ($-\\log[Pr(x)]$) associated with the observations (here, the observations include the inputs $u$ of the probabilistic network, but will also include the responses of the participant $y$ if there are some).\n",
@@ -1023,7 +1023,7 @@
" The decision rule is a function stating how the agent selects among all possible actions, given the state of the beliefs in the perceptual model, and optionally additional parameters. Programmatically, this is a Python function taking a perceptual model as input (i.e. an instance of the HGF class), and returning a sequence of actions. This can be used for simulation. The decision rule should be clearly defined in order to write the {term}`Response function`.\n",
"\n",
"Response function\n",
- " The response function is a term that we use specifically for this package ([pyhgf](https://github.com/ilabcode/pyhgf)). It refers to the Python function that, using a given HGF model and optional parameter, returns the surprise associated with the observed actions.\n",
+ " The response function is a term that we use specifically for this package ([pyhgf](https://github.com/ComputationalPsychiatry/pyhgf)). It refers to the Python function that, using a given HGF model and optional parameter, returns the surprise associated with the observed actions.\n",
"\n",
"```"
]
diff --git a/docs/source/notebooks/3-Multilevel_HGF.ipynb b/docs/source/notebooks/3-Multilevel_HGF.ipynb
index 691aea4eb..c7abd2603 100644
--- a/docs/source/notebooks/3-Multilevel_HGF.ipynb
+++ b/docs/source/notebooks/3-Multilevel_HGF.ipynb
@@ -32,7 +32,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/3-Multilevel_HGF.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/3-Multilevel_HGF.ipynb)"
]
},
{
@@ -137,7 +137,7 @@
"Luckily, we already have all the components in place to do that. We already used Bayesian networks in the previous sections when we were inferring the distribution of some parameters. Here, we only had one agent (i.e. one participant), and therefore did not need any hyperprior. We need to extend this approach a bit, and explicitly state that we want to fit many models (participants) simultaneously, and draw the values of some parameters from a hyper-prior (i.e. the group-level distribution).\n",
"\n",
"But before we move forward, maybe it is worth clarifying some of the terminology we use, especially as, starting from now, many things are called **networks** but are pointing to different parts of the workflow. We can indeed distinguish two kinds:\n",
- "1. The predictive coding neural networks. This is the kind of network that [pyhgf](https://github.com/ilabcode/pyhgf) is designed to handle (see {ref}`probabilistic_networks`). Every HGF model is an instance of such a network.\n",
+ "1. The predictive coding neural networks. This is the kind of network that [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) is designed to handle (see {ref}`probabilistic_networks`). Every HGF model is an instance of such a network.\n",
"2. The Bayesian (multilevel) network is the computational graph that is created with tools like [pymc](https://www.pymc.io/welcome.html). This graph will represent the dependencies between our variables and the way they are transformed.\n",
"\n",
"In this notebook, we are going to create the second type of network and incorporate many networks of the first type in it as custom distribution."
diff --git a/docs/source/notebooks/4-Parameter_recovery.ipynb b/docs/source/notebooks/4-Parameter_recovery.ipynb
index 1120471f0..f019c30d4 100644
--- a/docs/source/notebooks/4-Parameter_recovery.ipynb
+++ b/docs/source/notebooks/4-Parameter_recovery.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/4-Parameter_recovery.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/4-Parameter_recovery.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/5-Non_linear_value_coupling.ipynb b/docs/source/notebooks/5-Non_linear_value_coupling.ipynb
index 1daa5cdf7..cb78a2d8c 100644
--- a/docs/source/notebooks/5-Non_linear_value_coupling.ipynb
+++ b/docs/source/notebooks/5-Non_linear_value_coupling.ipynb
@@ -12,7 +12,7 @@
"cell_type": "markdown",
"metadata": {},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.4-Continuous_HGF_non_linear_value_coupling.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.4-Continuous_HGF_non_linear_value_coupling.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb b/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb
index 8ef988a6d..8078c3a78 100644
--- a/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb
+++ b/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb)"
]
},
{
@@ -106,7 +106,7 @@
"id": "48020213-7f26-4201-b4b9-f072231bc225",
"metadata": {},
"source": [
- "The nodalized version of the Hierarchical Gaussian Filter that is implemented in [pyhgf](https://github.com/ilabcode/pyhgf) opens the possibility to create filters with multiple inputs. Here, we illustrate how we can use this feature to create an agent that is filtering their physiological signals in real-time. We use a two-level Hierarchical Gaussian Filter to predict the dynamics of the instantaneous heart rate (the RR interval measured at each heartbeat). We then extract the trajectory of surprise at each predictive node to relate it with the cognitive task performed by the participant while the signal is being recorded."
+ "The nodalized version of the Hierarchical Gaussian Filter that is implemented in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) opens the possibility to create filters with multiple inputs. Here, we illustrate how we can use this feature to create an agent that is filtering their physiological signals in real-time. We use a two-level Hierarchical Gaussian Filter to predict the dynamics of the instantaneous heart rate (the RR interval measured at each heartbeat). We then extract the trajectory of surprise at each predictive node to relate it with the cognitive task performed by the participant while the signal is being recorded."
]
},
{
diff --git a/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb b/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb
index b11635c5a..5bbf09532 100644
--- a/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb
+++ b/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb b/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb
index 4ce529bfa..955bde00a 100644
--- a/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb
+++ b/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb b/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb
index 03c230d81..bc126d259 100644
--- a/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb
+++ b/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb)"
]
},
{
@@ -137,7 +137,7 @@
"x_1^{(k)} \\sim \\mathcal{N}(x_1^{(k-1)}, \\sigma^2)\n",
"$$\n",
"\n",
- "where $\\sigma^2$ is the fixed variance of the distribution. You can find more details on this as well as some code to get started with the simulations in the first tutorial on the [PyHGF documentation](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#).\n",
+ "where $\\sigma^2$ is the fixed variance of the distribution. You can find more details on this as well as some code to get started with the simulations in the first tutorial on the [PyHGF documentation](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#).\n",
"\n",
"```{exercise}\n",
":label: exercise1.1\n",
@@ -183,7 +183,7 @@
"$$\n",
"\n",
"```{hint}\n",
- "This generative process reads top-down: the node higher in the hierarchy ($x_2$) generates new values and passes them to the child nodes. We can also control how much the node $x_1$ is influenced by its previous (see examples [here](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#value-coupling)). To keep things simple in this exercise, we will assume that $x_1$ is fully influenced by its previous value, as noted in the equation, therefore $x_2$ can be seen as a *drift parent*.\n",
+ "This generative process reads top-down: the node higher in the hierarchy ($x_2$) generates new values and passes them to the child nodes. We can also control how much the node $x_1$ is influenced by its previous (see examples [here](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#value-coupling)). To keep things simple in this exercise, we will assume that $x_1$ is fully influenced by its previous value, as noted in the equation, therefore $x_2$ can be seen as a *drift parent*.\n",
"```\n",
"\n",
"Let's define a time series `x_2` that represent the evolution of the states of the parent node:"
@@ -373,11 +373,11 @@
"source": [
"Building on these principles, we can create networks of arbitrary size and shape made of probabilistic nodes (nodes that represent the evolution of a Gaussian Random Walk) that can be connected with each other through **value** or **volatility** coupling. We have covered these points in the previous exercise using a simplified network made of two nodes.\n",
"\n",
- "[PyHGF](https://ilabcode.github.io/pyhgf/index.html) adds most of its values in that it allows the creation and manipulation of large and complex networks easily, and it automates the transmission of values between nodes. You can refer to [the second part of the tutorial documentation](https://ilabcode.github.io/pyhgf/notebooks/0.2-Creating_networks.html) which covers the manipulation of networks for more details. \n",
+ "[PyHGF](https://computationalpsychiatry.github.io/pyhgf/index.html) adds most of its values in that it allows the creation and manipulation of large and complex networks easily, and it automates the transmission of values between nodes. You can refer to [the second part of the tutorial documentation](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.2-Creating_networks.html) which covers the manipulation of networks for more details. \n",
"\n",
"But this is not the only thing it does. So far we have simulated observations from the generative model, going for the leaves to the root of the network. This describes how we expect the environment to behave, but not how an agent would learn from it. To do so we need to invert this model: we want to provide the observations, and let the network update the nodes accordingly, so the beliefs always reflect the current state of the environment. This process requires propagating precision prediction errors from the root to the leaves of the network, and this is where most of PyHGF's dark magic is most useful.\n",
"\n",
- "```{figure} https://raw.githubusercontent.com/ilabcode/pyhgf/master/docs/source/images/graph_network.svg\n",
+ "```{figure} https://raw.githubusercontent.com/ComputationalPsychiatry/pyhgf/master/docs/source/images/graph_network.svg\n",
"---\n",
"name: networks\n",
"---\n",
@@ -760,7 +760,7 @@
],
"source": [
"aarhus_weather_df = pd.read_csv(\n",
- " \"https://raw.githubusercontent.com/ilabcode/hgf-data/main/datasets/weather.csv\"\n",
+ " \"https://raw.githubusercontent.com/ComputationalPsychiatry/hgf-data/main/datasets/weather.csv\"\n",
")\n",
"aarhus_weather_df.head()"
]
@@ -1076,7 +1076,7 @@
"````{solution} exercise1.5\n",
":label: solution-exercise1.5\n",
"\n",
- "This method return the Gaussian surprise at each time point, which are then summed. The sum of the Gaussian surprise reflect the performances of the model at predicting the next value, larger values pointing to a model more surprise by its inputs, therefore poor performances. The surprise is define as $s = -log(p)$, this is thus the negative of the log probability function. Log probability functions are commonly used by sampling algorithm, it is thus straigthforwars to sample a model parameters when this function is available. There are an infinity of response possible functions - just like there is an infinity of possible networks - for more details on how to use tem, you can refer to the [tutorial on custom response models](https://ilabcode.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html).\n",
+ "This method return the Gaussian surprise at each time point, which are then summed. The sum of the Gaussian surprise reflect the performances of the model at predicting the next value, larger values pointing to a model more surprise by its inputs, therefore poor performances. The surprise is define as $s = -log(p)$, this is thus the negative of the log probability function. Log probability functions are commonly used by sampling algorithm, it is thus straigthforwars to sample a model parameters when this function is available. There are an infinity of response possible functions - just like there is an infinity of possible networks - for more details on how to use tem, you can refer to the [tutorial on custom response models](https://computationalpsychiatry.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html).\n",
"\n",
"````"
]
diff --git a/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb b/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
index 7a1b0f4c1..0dc3ce8c6 100644
--- a/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
+++ b/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb)"
]
},
{
@@ -1942,9 +1942,9 @@
"\n",
"All these sections should give you a solid understanding of the model and how to use it in context. If you want to apply this to your dataset, we recommend exploring the tutorial section of the documentation:\n",
"\n",
- "- [Using custom response models](https://ilabcode.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html) will explore the creation of custom models for behaviours that can match your experimental design and theory.\n",
- "- [Multilevel modelling](https://ilabcode.github.io/pyhgf/notebooks/3-Multilevel_HGF.html) will discuss modelling at the group/condition level in multilevel Bayesian networks.\n",
- "- [Parameter recovery](https://ilabcode.github.io/pyhgf/notebooks/4-Parameter_recovery.html) explains how to simulate a dataset and perform parameter recovery, as a prior validation of your models."
+ "- [Using custom response models](https://computationalpsychiatry.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html) will explore the creation of custom models for behaviours that can match your experimental design and theory.\n",
+ "- [Multilevel modelling](https://computationalpsychiatry.github.io/pyhgf/notebooks/3-Multilevel_HGF.html) will discuss modelling at the group/condition level in multilevel Bayesian networks.\n",
+ "- [Parameter recovery](https://computationalpsychiatry.github.io/pyhgf/notebooks/4-Parameter_recovery.html) explains how to simulate a dataset and perform parameter recovery, as a prior validation of your models."
]
},
{
diff --git a/pyproject.toml b/pyproject.toml
index 678d407fd..d9a4246d3 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -4,8 +4,8 @@ description = "Dynamic neural networks for predictive coding"
authors = ["Nicolas Legrand "]
license = "GPL-3.0"
readme = "README.md"
-homepage = "https://ilabcode.github.io/pyhgf/"
-repository = "https://github.com/ilabcode/pyhgf"
+homepage = "https://computationalpsychiatry.github.io/pyhgf/"
+repository = "https://github.com/ComputationalPsychiatry/pyhgf"
keywords = ["reinforcement learning", "predictive coding", "neural networks", "graphs", "variational inference", "active inference", "causal inference"]
include = [
"src/pyhgf/data/usdchf.txt",
You can find a deeper introduction to how to create and manipulate networks under the following link:
-* 🎓 [Creating and manipulating networks of probabilistic nodes](https://ilabcode.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
+* 🎓 [Creating and manipulating networks of probabilistic nodes](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
### The Generalized Hierarchical Gaussian Filter
@@ -49,7 +49,7 @@ Generalized Hierarchical Gaussian Filters (gHGF) are specific instances of dynam
You can find a deeper introduction on how does the gHGF works under the following link:
-* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
+* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
### Model fitting
@@ -76,7 +76,7 @@ hgf.input_data(input_data=u)
hgf.plot_trajectories();
```
-
+
```python
from pyhgf.response import binary_softmax_inverse_temperature
@@ -96,7 +96,7 @@ print(f"Sum of surprises = {surprise.sum()}")
## Acknowledgments
-This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ilabcode/HGF.jl).
+This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ComputationalPsychiatry/HGF.jl).
## References
diff --git a/docs/source/learn.md b/docs/source/learn.md
index f99e8672c..076e1f8cb 100644
--- a/docs/source/learn.md
+++ b/docs/source/learn.md
@@ -50,7 +50,7 @@ notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
# Learn
-In this section, you can find tutorial notebooks that describe the internals of pyhgf, the theory behind the Hierarchical Gaussian filter, and step-by-step application and use cases of the model. At the beginning of every tutorial, you will find a badge [](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.1-Creating_networks.ipynb) to run the notebook interactively in a Google Colab session.
+In this section, you can find tutorial notebooks that describe the internals of pyhgf, the theory behind the Hierarchical Gaussian filter, and step-by-step application and use cases of the model. At the beginning of every tutorial, you will find a badge [](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.1-Creating_networks.ipynb) to run the notebook interactively in a Google Colab session.
## Theory
diff --git a/docs/source/notebooks/0.1-Theory.ipynb b/docs/source/notebooks/0.1-Theory.ipynb
index c995adf49..af2327fbe 100644
--- a/docs/source/notebooks/0.1-Theory.ipynb
+++ b/docs/source/notebooks/0.1-Theory.ipynb
@@ -14,7 +14,7 @@
"(theory)=\n",
"# Introduction to the Generalised Hierarchical Gaussian Filter\n",
"\n",
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.1-Theory.ipynb)\n",
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.1-Theory.ipynb)\n",
"\n",
"The Hierarchical Gaussian Filter is a Bayesian model of belief updating under uncertainty in which the volatility of the environment is encoded by a hierarchy of probability distributions. A generalised version of this model {cite:p}`weber:2023` has further framed this into a neural network framework of probabilistic state nodes interacting with each other. At the heart of both frameworks lies a generative model that consist in a hierarchy of {term}`Gaussian Random Walk`. The inversion of this model (i.e. estimating parameters from observed values) leads to the derivation of prediction errors and posterior updates that can turn these networks into learning systems. To fully understand this model, it is therefore central to start with the simplest case, the implied generative model, which can be seen as the probabilistic structure that generates observations.\n",
"\n",
@@ -664,7 +664,7 @@
"source": [
"### The propagation of prediction and prediction errors\n",
"\n",
- "Having described the model as a specific configuration of predictive nodes offer many advantages, especially in term of modularity for the user. However, the model itself is not limited to the description of the generative process that we covered in the previous examples. The most interesting, and also the more complex, part of the modelling consists of the capability for the network to update the hierarchical structure in a Bayesian optimal way as new observations are presented. These steps are defined by a set of simple, one-step update equations that represent changes in beliefs about the hidden states (i.e. the sufficient statistics of the nodes) specified in the generative model. These equations were first described in {cite:t}`2011:mathys`, and the update equations for volatility and value coupling in the generalized Hierarchical Gaussian filter (on which most of the update functions in [pyhgf](https://github.com/ilabcode/pyhgf) are based) have been described in {cite:p}`weber:2023`. The exact computations in each step especially depend on the nature of the coupling (via {term}`VAPE`s vs. {term}`VOPE`s) between the parent and children nodes. It is beyond the scope of this tutorial to dive into the derivation of these steps and we refer the interested reader to the mentioned papers. Here, we provide a general overview of the dynamic of the update sequence that supports belief updating. The computations triggered by any observation at each time point can be ordered in time as shown in the belief update algorithm."
+ "Having described the model as a specific configuration of predictive nodes offer many advantages, especially in term of modularity for the user. However, the model itself is not limited to the description of the generative process that we covered in the previous examples. The most interesting, and also the more complex, part of the modelling consists of the capability for the network to update the hierarchical structure in a Bayesian optimal way as new observations are presented. These steps are defined by a set of simple, one-step update equations that represent changes in beliefs about the hidden states (i.e. the sufficient statistics of the nodes) specified in the generative model. These equations were first described in {cite:t}`2011:mathys`, and the update equations for volatility and value coupling in the generalized Hierarchical Gaussian filter (on which most of the update functions in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) are based) have been described in {cite:p}`weber:2023`. The exact computations in each step especially depend on the nature of the coupling (via {term}`VAPE`s vs. {term}`VOPE`s) between the parent and children nodes. It is beyond the scope of this tutorial to dive into the derivation of these steps and we refer the interested reader to the mentioned papers. Here, we provide a general overview of the dynamic of the update sequence that supports belief updating. The computations triggered by any observation at each time point can be ordered in time as shown in the belief update algorithm."
]
},
{
diff --git a/docs/source/notebooks/0.2-Creating_networks.ipynb b/docs/source/notebooks/0.2-Creating_networks.ipynb
index 82dffc43c..c3640e0e5 100644
--- a/docs/source/notebooks/0.2-Creating_networks.ipynb
+++ b/docs/source/notebooks/0.2-Creating_networks.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.2-Creating_networks.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.2-Creating_networks.ipynb)"
]
},
{
@@ -85,7 +85,7 @@
"id": "57dde3f9-d8b1-437e-8a33-7aea1a1b1e2e",
"metadata": {},
"source": [
- "[pyhgf](https://ilabcode.github.io/pyhgf/index.html#) is designed with inspiration from graph neural network libraries that can support message-passing schemes and perform belief propagation through networks of probabilistic nodes. Here, this principle is applied to predictive processing and focuses on networks that are structured as **rooted trees** and perform variational message passing to update beliefs about the state of the environment, inferred from the observations at the root of the tree. While this library is optimized to implement the standard two-level and three-level HGF {cite:p}`2011:mathys,2014:mathys`, as well as the generalized HGF {cite:p}`weber:2023`, it can also be applied to much larger use cases, with the idea is to generalize belief propagation as it has been described so far to larger and more complex networks that will capture a greater variety of environmental structure. Therefore, the library is also designed to facilitate the creation and manipulation of such probabilistic networks. Importantly, here we consider that a probabilistic network should be defined by the following four variables:\n",
+ "[pyhgf](https://computationalpsychiatry.github.io/pyhgf/index.html#) is designed with inspiration from graph neural network libraries that can support message-passing schemes and perform belief propagation through networks of probabilistic nodes. Here, this principle is applied to predictive processing and focuses on networks that are structured as **rooted trees** and perform variational message passing to update beliefs about the state of the environment, inferred from the observations at the root of the tree. While this library is optimized to implement the standard two-level and three-level HGF {cite:p}`2011:mathys,2014:mathys`, as well as the generalized HGF {cite:p}`weber:2023`, it can also be applied to much larger use cases, with the idea is to generalize belief propagation as it has been described so far to larger and more complex networks that will capture a greater variety of environmental structure. Therefore, the library is also designed to facilitate the creation and manipulation of such probabilistic networks. Importantly, here we consider that a probabilistic network should be defined by the following four variables:\n",
"1. the network parameters\n",
"2. the network structure\n",
"3. the update function(s)\n",
@@ -711,7 +711,7 @@
"source": [
"### Multivariate coupling\n",
"\n",
- "As we can see in the examples above, nodes in a valid HGF network can be influenced by multiple parents (either value or volatility parents). Similarly, a single node can be influenced by multiple children. This feature is termed *multivariate descendency* and *multivariate ascendency* (respectively) and is a central addition to the generalization of the HGF {cite:p}`weber:2023` that was implemented in this package, as well as in the [Julia counterpart](https://github.com/ilabcode/HierarchicalGaussianFiltering.jl)."
+ "As we can see in the examples above, nodes in a valid HGF network can be influenced by multiple parents (either value or volatility parents). Similarly, a single node can be influenced by multiple children. This feature is termed *multivariate descendency* and *multivariate ascendency* (respectively) and is a central addition to the generalization of the HGF {cite:p}`weber:2023` that was implemented in this package, as well as in the [Julia counterpart](https://github.com/ComputationalPsychiatry/HierarchicalGaussianFiltering.jl)."
]
},
{
@@ -1748,7 +1748,7 @@
"source": [
"## Creating custom update functions\n",
"\n",
- "The structure of the network and the node's parameters are the most static component of the network. Actually, we could consider that the network already exists once those two variables are in place. However in [pyhgf](https://ilabcode.github.io/pyhgf/index.html#) we consider that the update functions $\\mathcal{F} = \\{f_1, ..., f_n\\}$ and the update sequence $\\Sigma = [f_1(n_1), ..., f_i(n_j), f \\in \\mathcal{F}, n \\in 1, ..., k ]$ (the order of update) are also part of the models. This choice was made to explicitly account that there is no one unique way of modelling the way beliefs propagate through the network, and a core task for predictive coding applications is to develop new *probabilistic nodes* that account for a greater variety of phenomena. This step critically requires modelling beliefs diffusion, and therefore to modify, or creating the underlying update functions."
+ "The structure of the network and the node's parameters are the most static component of the network. Actually, we could consider that the network already exists once those two variables are in place. However in [pyhgf](https://computationalpsychiatry.github.io/pyhgf/index.html#) we consider that the update functions $\\mathcal{F} = \\{f_1, ..., f_n\\}$ and the update sequence $\\Sigma = [f_1(n_1), ..., f_i(n_j), f \\in \\mathcal{F}, n \\in 1, ..., k ]$ (the order of update) are also part of the models. This choice was made to explicitly account that there is no one unique way of modelling the way beliefs propagate through the network, and a core task for predictive coding applications is to develop new *probabilistic nodes* that account for a greater variety of phenomena. This step critically requires modelling beliefs diffusion, and therefore to modify, or creating the underlying update functions."
]
},
{
diff --git a/docs/source/notebooks/0.3-Generalised_filtering.ipynb b/docs/source/notebooks/0.3-Generalised_filtering.ipynb
index 552989459..5452eb9ce 100644
--- a/docs/source/notebooks/0.3-Generalised_filtering.ipynb
+++ b/docs/source/notebooks/0.3-Generalised_filtering.ipynb
@@ -14,7 +14,7 @@
"(generalised_filtering)=\n",
"# From Reinforcement Learning to Generalised Bayesian Filtering\n",
"\n",
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.3-Generalised_filtering.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.3-Generalised_filtering.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/1.1-Binary_HGF.ipynb b/docs/source/notebooks/1.1-Binary_HGF.ipynb
index 83297bccc..5d02d4deb 100644
--- a/docs/source/notebooks/1.1-Binary_HGF.ipynb
+++ b/docs/source/notebooks/1.1-Binary_HGF.ipynb
@@ -20,7 +20,7 @@
"id": "1258ea53-11fc-4a27-927c-2afc91f4aa12",
"metadata": {},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.1-Binary_HGF.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.1-Binary_HGF.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/1.2-Categorical_HGF.ipynb b/docs/source/notebooks/1.2-Categorical_HGF.ipynb
index f4a42850f..06ab288d2 100644
--- a/docs/source/notebooks/1.2-Categorical_HGF.ipynb
+++ b/docs/source/notebooks/1.2-Categorical_HGF.ipynb
@@ -20,7 +20,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.3-CAtegorical_HGF.ipynb)\n",
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.3-CAtegorical_HGF.ipynb)\n",
"\n",
"```{warning}\n",
"The categorical state node and the categorical state-transition nodes are still work in progress. The examples provided here are given for illustration. Things may change or not work until the official publication.\n",
diff --git a/docs/source/notebooks/1.3-Continuous_HGF.ipynb b/docs/source/notebooks/1.3-Continuous_HGF.ipynb
index 84e0def7e..db1e94681 100644
--- a/docs/source/notebooks/1.3-Continuous_HGF.ipynb
+++ b/docs/source/notebooks/1.3-Continuous_HGF.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.3-Continuous_HGF.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.3-Continuous_HGF.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/2-Using_custom_response_functions.ipynb b/docs/source/notebooks/2-Using_custom_response_functions.ipynb
index 71554bcb4..774e43dad 100644
--- a/docs/source/notebooks/2-Using_custom_response_functions.ipynb
+++ b/docs/source/notebooks/2-Using_custom_response_functions.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/2-Using_custom_response_functions.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/2-Using_custom_response_functions.ipynb)"
]
},
{
@@ -542,12 +542,12 @@
"\n",
"Let's now consider that the two vectors of observations $u$ and responses $y$ were obtained from a real participant undergoing a real experiment. In this situation, we assume that this participant internally used a {term}`Perceptual model` and a {term}`Decision rule` that might resemble what we defined previously, and we want to infer what are the most likely values for critical parameters in the model (e.g. the evolution rate $\\omega_2$). To do so, we are going to use our dataset (both $u$ and $y$), and try many models. We are going to fix the values of all HGF parameters to reasonable estimates (here, using the exact same values as in the simulation), except for $\\omega_2$. For this last parameter, we will assume a prior set at $\\mathcal{N}(-2.0, 2.0)$. The idea is that we want to sample many $\\omega_2$ values from this distribution and see if the model is performing better with some values.\n",
"\n",
- "But here, we need a clear definition of what this means *to perform better* for a given model. And this is exactly what a {term}`Response model` does, it is a way for us to evaluate how likely the behaviours $y$ for a given {term}`Perceptual model`, and assuming that the participants use this specific {term}`Decision rule`. In [pyhgf](https://github.com/ilabcode/pyhgf), this step is performed by creating the corresponding {term}`Response function`, which is the Python function that will return the surprise $S$ of getting these behaviours from the participant under this decision rule.\n",
+ "But here, we need a clear definition of what this means *to perform better* for a given model. And this is exactly what a {term}`Response model` does, it is a way for us to evaluate how likely the behaviours $y$ for a given {term}`Perceptual model`, and assuming that the participants use this specific {term}`Decision rule`. In [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), this step is performed by creating the corresponding {term}`Response function`, which is the Python function that will return the surprise $S$ of getting these behaviours from the participant under this decision rule.\n",
"\n",
"````{hint} What is a *response function*?\n",
"Most of the work around HGFs consists in creating and adapting {term}`Response model` to work with a given experimental design. There is no limit in terms of what can be used as a {term}`Response model`, provided that the {term}`Perceptual model` and the {term}`Decision rule` are clearly defined.\n",
"\n",
- "In [pyhgf](https://github.com/ilabcode/pyhgf), the {term}`Perceptual model` is the probabilistic network created with the main {py:class}`pyhgf.model.HGF` and {py:class}`pyhgf.distribution.HGFDistribution` classes. The {term}`Response model` is something that is implicitly defined when we create the {term}`Response function`, a Python function that computes the negative of the log-likelihood of the actions given the perceptual model. This {term}`Response function` can be passed as an argument to the main classes using the keywords arguments `response_function`, `response_function_inputs` and `response_function_parameters`. The `response_function` can be any callable that returns the surprise $S$ of observing action $y$ given this model, and the {term}`Decision rule`. The `response_function_inputs` are the additional data to the response function (optional) while `response_function_parameters` are the additional parameters (optional). The `response_function_inputs` is where the actions $y$ should be provided.\n",
+ "In [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), the {term}`Perceptual model` is the probabilistic network created with the main {py:class}`pyhgf.model.HGF` and {py:class}`pyhgf.distribution.HGFDistribution` classes. The {term}`Response model` is something that is implicitly defined when we create the {term}`Response function`, a Python function that computes the negative of the log-likelihood of the actions given the perceptual model. This {term}`Response function` can be passed as an argument to the main classes using the keywords arguments `response_function`, `response_function_inputs` and `response_function_parameters`. The `response_function` can be any callable that returns the surprise $S$ of observing action $y$ given this model, and the {term}`Decision rule`. The `response_function_inputs` are the additional data to the response function (optional) while `response_function_parameters` are the additional parameters (optional). The `response_function_inputs` is where the actions $y$ should be provided.\n",
"\n",
"```{important}\n",
"A *response function* should not return the actions given perceptual inputs $y | u$ (this is what the {term}`Decision rule` does), but the [surprise](https://en.wikipedia.org/wiki/Information_content) $S$ associated with the observation of actions given the perceptual inputs $S(y | u)$, which is defined by:\n",
@@ -559,7 +559,7 @@
"$$\n",
"```\n",
"\n",
- "If you are already familiar with using HGFs in the Julia equivalent of [pyhgf](https://github.com/ilabcode/pyhgf), you probably noted that the toolbox is split into a **perceptual** package [HierarchicalGaussianFiltering.jl](https://github.com/ilabcode/HierarchicalGaussianFiltering.jl) and a **response** package [ActionModels.jl](https://github.com/ilabcode/ActionModels.jl). This was made to make the difference between the two parts of the HGF clear and be explicit that you can use a perceptual model without any action model. In [pyhgf](https://github.com/ilabcode/pyhgf) however, everything happens in the same package, the response function is merely an optional, additional argument that can be passed to describe how surprise is computed.\n",
+ "If you are already familiar with using HGFs in the Julia equivalent of [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), you probably noted that the toolbox is split into a **perceptual** package [HierarchicalGaussianFiltering.jl](https://github.com/ComputationalPsychiatry/HierarchicalGaussianFiltering.jl) and a **response** package [ActionModels.jl](https://github.com/ComputationalPsychiatry/ActionModels.jl). This was made to make the difference between the two parts of the HGF clear and be explicit that you can use a perceptual model without any action model. In [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) however, everything happens in the same package, the response function is merely an optional, additional argument that can be passed to describe how surprise is computed.\n",
"````"
]
},
@@ -637,7 +637,7 @@
},
"source": [
"```{note}\n",
- "Note here that our {term}`Response function` has a structure that is the standard way to write response functions in [pyhgf](https://github.com/ilabcode/pyhgf), that is with two input arguments:\n",
+ "Note here that our {term}`Response function` has a structure that is the standard way to write response functions in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), that is with two input arguments:\n",
"- the HGF model on which the response function applies (i.e. the {term}`Perceptual model`)\n",
"- the additional parameters provided to the response function. This can include additional parameters that can be part of the equation of the model, or the input data used by this model. We then provide the `response` vector ($y$) here.\n",
"\n",
@@ -1014,7 +1014,7 @@
"```{glossary}\n",
"\n",
"Perceptual model\n",
- " The perceptual model of a Hierarchical Gaussian Filter traditionally refers to the branch receiving observations $u$ about states of the world and that performs the updating of beliefs about these states. By generalisation, the perceptual model is any probabilistic network that can be created in [pyhgf](https://github.com/ilabcode/pyhgf), receiving an arbitrary number of inputs. An HGF that only consists of a perceptual model will act as a Bayesian filter.\n",
+ " The perceptual model of a Hierarchical Gaussian Filter traditionally refers to the branch receiving observations $u$ about states of the world and that performs the updating of beliefs about these states. By generalisation, the perceptual model is any probabilistic network that can be created in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), receiving an arbitrary number of inputs. An HGF that only consists of a perceptual model will act as a Bayesian filter.\n",
"\n",
"Response model\n",
" The response model of a Hierarchical Gaussian filter refers to the branch that uses the beliefs about the state of the world to generate actions using the {term}`Decision rule`. This branch is also sometimes referred to as the **decision model** or the **observation model**, depending on the fields. Critically, this part of the model can return the surprise ($-\\log[Pr(x)]$) associated with the observations (here, the observations include the inputs $u$ of the probabilistic network, but will also include the responses of the participant $y$ if there are some).\n",
@@ -1023,7 +1023,7 @@
" The decision rule is a function stating how the agent selects among all possible actions, given the state of the beliefs in the perceptual model, and optionally additional parameters. Programmatically, this is a Python function taking a perceptual model as input (i.e. an instance of the HGF class), and returning a sequence of actions. This can be used for simulation. The decision rule should be clearly defined in order to write the {term}`Response function`.\n",
"\n",
"Response function\n",
- " The response function is a term that we use specifically for this package ([pyhgf](https://github.com/ilabcode/pyhgf)). It refers to the Python function that, using a given HGF model and optional parameter, returns the surprise associated with the observed actions.\n",
+ " The response function is a term that we use specifically for this package ([pyhgf](https://github.com/ComputationalPsychiatry/pyhgf)). It refers to the Python function that, using a given HGF model and optional parameter, returns the surprise associated with the observed actions.\n",
"\n",
"```"
]
diff --git a/docs/source/notebooks/3-Multilevel_HGF.ipynb b/docs/source/notebooks/3-Multilevel_HGF.ipynb
index 691aea4eb..c7abd2603 100644
--- a/docs/source/notebooks/3-Multilevel_HGF.ipynb
+++ b/docs/source/notebooks/3-Multilevel_HGF.ipynb
@@ -32,7 +32,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/3-Multilevel_HGF.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/3-Multilevel_HGF.ipynb)"
]
},
{
@@ -137,7 +137,7 @@
"Luckily, we already have all the components in place to do that. We already used Bayesian networks in the previous sections when we were inferring the distribution of some parameters. Here, we only had one agent (i.e. one participant), and therefore did not need any hyperprior. We need to extend this approach a bit, and explicitly state that we want to fit many models (participants) simultaneously, and draw the values of some parameters from a hyper-prior (i.e. the group-level distribution).\n",
"\n",
"But before we move forward, maybe it is worth clarifying some of the terminology we use, especially as, starting from now, many things are called **networks** but are pointing to different parts of the workflow. We can indeed distinguish two kinds:\n",
- "1. The predictive coding neural networks. This is the kind of network that [pyhgf](https://github.com/ilabcode/pyhgf) is designed to handle (see {ref}`probabilistic_networks`). Every HGF model is an instance of such a network.\n",
+ "1. The predictive coding neural networks. This is the kind of network that [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) is designed to handle (see {ref}`probabilistic_networks`). Every HGF model is an instance of such a network.\n",
"2. The Bayesian (multilevel) network is the computational graph that is created with tools like [pymc](https://www.pymc.io/welcome.html). This graph will represent the dependencies between our variables and the way they are transformed.\n",
"\n",
"In this notebook, we are going to create the second type of network and incorporate many networks of the first type in it as custom distribution."
diff --git a/docs/source/notebooks/4-Parameter_recovery.ipynb b/docs/source/notebooks/4-Parameter_recovery.ipynb
index 1120471f0..f019c30d4 100644
--- a/docs/source/notebooks/4-Parameter_recovery.ipynb
+++ b/docs/source/notebooks/4-Parameter_recovery.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/4-Parameter_recovery.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/4-Parameter_recovery.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/5-Non_linear_value_coupling.ipynb b/docs/source/notebooks/5-Non_linear_value_coupling.ipynb
index 1daa5cdf7..cb78a2d8c 100644
--- a/docs/source/notebooks/5-Non_linear_value_coupling.ipynb
+++ b/docs/source/notebooks/5-Non_linear_value_coupling.ipynb
@@ -12,7 +12,7 @@
"cell_type": "markdown",
"metadata": {},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.4-Continuous_HGF_non_linear_value_coupling.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.4-Continuous_HGF_non_linear_value_coupling.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb b/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb
index 8ef988a6d..8078c3a78 100644
--- a/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb
+++ b/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb)"
]
},
{
@@ -106,7 +106,7 @@
"id": "48020213-7f26-4201-b4b9-f072231bc225",
"metadata": {},
"source": [
- "The nodalized version of the Hierarchical Gaussian Filter that is implemented in [pyhgf](https://github.com/ilabcode/pyhgf) opens the possibility to create filters with multiple inputs. Here, we illustrate how we can use this feature to create an agent that is filtering their physiological signals in real-time. We use a two-level Hierarchical Gaussian Filter to predict the dynamics of the instantaneous heart rate (the RR interval measured at each heartbeat). We then extract the trajectory of surprise at each predictive node to relate it with the cognitive task performed by the participant while the signal is being recorded."
+ "The nodalized version of the Hierarchical Gaussian Filter that is implemented in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) opens the possibility to create filters with multiple inputs. Here, we illustrate how we can use this feature to create an agent that is filtering their physiological signals in real-time. We use a two-level Hierarchical Gaussian Filter to predict the dynamics of the instantaneous heart rate (the RR interval measured at each heartbeat). We then extract the trajectory of surprise at each predictive node to relate it with the cognitive task performed by the participant while the signal is being recorded."
]
},
{
diff --git a/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb b/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb
index b11635c5a..5bbf09532 100644
--- a/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb
+++ b/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb b/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb
index 4ce529bfa..955bde00a 100644
--- a/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb
+++ b/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb b/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb
index 03c230d81..bc126d259 100644
--- a/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb
+++ b/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb)"
]
},
{
@@ -137,7 +137,7 @@
"x_1^{(k)} \\sim \\mathcal{N}(x_1^{(k-1)}, \\sigma^2)\n",
"$$\n",
"\n",
- "where $\\sigma^2$ is the fixed variance of the distribution. You can find more details on this as well as some code to get started with the simulations in the first tutorial on the [PyHGF documentation](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#).\n",
+ "where $\\sigma^2$ is the fixed variance of the distribution. You can find more details on this as well as some code to get started with the simulations in the first tutorial on the [PyHGF documentation](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#).\n",
"\n",
"```{exercise}\n",
":label: exercise1.1\n",
@@ -183,7 +183,7 @@
"$$\n",
"\n",
"```{hint}\n",
- "This generative process reads top-down: the node higher in the hierarchy ($x_2$) generates new values and passes them to the child nodes. We can also control how much the node $x_1$ is influenced by its previous (see examples [here](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#value-coupling)). To keep things simple in this exercise, we will assume that $x_1$ is fully influenced by its previous value, as noted in the equation, therefore $x_2$ can be seen as a *drift parent*.\n",
+ "This generative process reads top-down: the node higher in the hierarchy ($x_2$) generates new values and passes them to the child nodes. We can also control how much the node $x_1$ is influenced by its previous (see examples [here](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#value-coupling)). To keep things simple in this exercise, we will assume that $x_1$ is fully influenced by its previous value, as noted in the equation, therefore $x_2$ can be seen as a *drift parent*.\n",
"```\n",
"\n",
"Let's define a time series `x_2` that represent the evolution of the states of the parent node:"
@@ -373,11 +373,11 @@
"source": [
"Building on these principles, we can create networks of arbitrary size and shape made of probabilistic nodes (nodes that represent the evolution of a Gaussian Random Walk) that can be connected with each other through **value** or **volatility** coupling. We have covered these points in the previous exercise using a simplified network made of two nodes.\n",
"\n",
- "[PyHGF](https://ilabcode.github.io/pyhgf/index.html) adds most of its values in that it allows the creation and manipulation of large and complex networks easily, and it automates the transmission of values between nodes. You can refer to [the second part of the tutorial documentation](https://ilabcode.github.io/pyhgf/notebooks/0.2-Creating_networks.html) which covers the manipulation of networks for more details. \n",
+ "[PyHGF](https://computationalpsychiatry.github.io/pyhgf/index.html) adds most of its values in that it allows the creation and manipulation of large and complex networks easily, and it automates the transmission of values between nodes. You can refer to [the second part of the tutorial documentation](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.2-Creating_networks.html) which covers the manipulation of networks for more details. \n",
"\n",
"But this is not the only thing it does. So far we have simulated observations from the generative model, going for the leaves to the root of the network. This describes how we expect the environment to behave, but not how an agent would learn from it. To do so we need to invert this model: we want to provide the observations, and let the network update the nodes accordingly, so the beliefs always reflect the current state of the environment. This process requires propagating precision prediction errors from the root to the leaves of the network, and this is where most of PyHGF's dark magic is most useful.\n",
"\n",
- "```{figure} https://raw.githubusercontent.com/ilabcode/pyhgf/master/docs/source/images/graph_network.svg\n",
+ "```{figure} https://raw.githubusercontent.com/ComputationalPsychiatry/pyhgf/master/docs/source/images/graph_network.svg\n",
"---\n",
"name: networks\n",
"---\n",
@@ -760,7 +760,7 @@
],
"source": [
"aarhus_weather_df = pd.read_csv(\n",
- " \"https://raw.githubusercontent.com/ilabcode/hgf-data/main/datasets/weather.csv\"\n",
+ " \"https://raw.githubusercontent.com/ComputationalPsychiatry/hgf-data/main/datasets/weather.csv\"\n",
")\n",
"aarhus_weather_df.head()"
]
@@ -1076,7 +1076,7 @@
"````{solution} exercise1.5\n",
":label: solution-exercise1.5\n",
"\n",
- "This method return the Gaussian surprise at each time point, which are then summed. The sum of the Gaussian surprise reflect the performances of the model at predicting the next value, larger values pointing to a model more surprise by its inputs, therefore poor performances. The surprise is define as $s = -log(p)$, this is thus the negative of the log probability function. Log probability functions are commonly used by sampling algorithm, it is thus straigthforwars to sample a model parameters when this function is available. There are an infinity of response possible functions - just like there is an infinity of possible networks - for more details on how to use tem, you can refer to the [tutorial on custom response models](https://ilabcode.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html).\n",
+ "This method return the Gaussian surprise at each time point, which are then summed. The sum of the Gaussian surprise reflect the performances of the model at predicting the next value, larger values pointing to a model more surprise by its inputs, therefore poor performances. The surprise is define as $s = -log(p)$, this is thus the negative of the log probability function. Log probability functions are commonly used by sampling algorithm, it is thus straigthforwars to sample a model parameters when this function is available. There are an infinity of response possible functions - just like there is an infinity of possible networks - for more details on how to use tem, you can refer to the [tutorial on custom response models](https://computationalpsychiatry.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html).\n",
"\n",
"````"
]
diff --git a/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb b/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
index 7a1b0f4c1..0dc3ce8c6 100644
--- a/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
+++ b/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb)"
]
},
{
@@ -1942,9 +1942,9 @@
"\n",
"All these sections should give you a solid understanding of the model and how to use it in context. If you want to apply this to your dataset, we recommend exploring the tutorial section of the documentation:\n",
"\n",
- "- [Using custom response models](https://ilabcode.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html) will explore the creation of custom models for behaviours that can match your experimental design and theory.\n",
- "- [Multilevel modelling](https://ilabcode.github.io/pyhgf/notebooks/3-Multilevel_HGF.html) will discuss modelling at the group/condition level in multilevel Bayesian networks.\n",
- "- [Parameter recovery](https://ilabcode.github.io/pyhgf/notebooks/4-Parameter_recovery.html) explains how to simulate a dataset and perform parameter recovery, as a prior validation of your models."
+ "- [Using custom response models](https://computationalpsychiatry.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html) will explore the creation of custom models for behaviours that can match your experimental design and theory.\n",
+ "- [Multilevel modelling](https://computationalpsychiatry.github.io/pyhgf/notebooks/3-Multilevel_HGF.html) will discuss modelling at the group/condition level in multilevel Bayesian networks.\n",
+ "- [Parameter recovery](https://computationalpsychiatry.github.io/pyhgf/notebooks/4-Parameter_recovery.html) explains how to simulate a dataset and perform parameter recovery, as a prior validation of your models."
]
},
{
diff --git a/pyproject.toml b/pyproject.toml
index 678d407fd..d9a4246d3 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -4,8 +4,8 @@ description = "Dynamic neural networks for predictive coding"
authors = ["Nicolas Legrand "]
license = "GPL-3.0"
readme = "README.md"
-homepage = "https://ilabcode.github.io/pyhgf/"
-repository = "https://github.com/ilabcode/pyhgf"
+homepage = "https://computationalpsychiatry.github.io/pyhgf/"
+repository = "https://github.com/ComputationalPsychiatry/pyhgf"
keywords = ["reinforcement learning", "predictive coding", "neural networks", "graphs", "variational inference", "active inference", "causal inference"]
include = [
"src/pyhgf/data/usdchf.txt",
 +
+ PyHGF is a Python library for creating and manipulating dynamic probabilistic networks for predictive coding. These networks approximate Bayesian inference by optimizing beliefs through the diffusion of predictions and precision-weighted prediction errors. The network structure remains flexible during message-passing steps, allowing for dynamic adjustments. They can be used as a biologically plausible cognitive model in computational neuroscience or as a generalization of Bayesian filtering for designing efficient, modular decision-making agents. The default implementation supports the generalized Hierarchical Gaussian Filters (gHGF, Weber et al., 2024), but the framework is designed to be adaptable to other algorithms. Built on top of JAX, the core functions are differentiable and JIT-compiled where applicable. The library is optimized for modularity and ease of use, allowing seamless integration with other libraries in the ecosystem for Bayesian inference and optimization. Additionally, a binding with an implementation in Rust is under active development, which will further enhance flexibility during inference. You can find the method paper describing the toolbox [here](https://arxiv.org/abs/2410.09206) and the method paper describing the gHGF, which is the main framework currently supported by the toolbox [here](https://arxiv.org/abs/2305.10937).
-* 📖 [API Documentation](https://ilabcode.github.io/pyhgf/api.html)
-* ✏️ [Tutorials and examples](https://ilabcode.github.io/pyhgf/learn.html)
+* 📖 [API Documentation](https://computationalpsychiatry.github.io/pyhgf/api.html)
+* ✏️ [Tutorials and examples](https://computationalpsychiatryionalpsychiatry.github.io/pyhgf/learn.html)
## Getting started
@@ -21,7 +21,7 @@ The last official release can be downloaded from PIP:
The current version under development can be installed from the master branch of the GitHub folder:
-`pip install “git+https://github.com/ilabcode/pyhgf.git”`
+`pip install “git+https://github.com/ComputationalPsychiatry/pyhgf.git”`
### How does it work?
@@ -32,12 +32,12 @@ Dynamic networks can be defined as a tuple containing the following variables:
* A set of update functions. An update function receive a network tuple and returns an updated network tuple.
* An update sequence (tuple) that defines the order and target of the update functions.
-
PyHGF is a Python library for creating and manipulating dynamic probabilistic networks for predictive coding. These networks approximate Bayesian inference by optimizing beliefs through the diffusion of predictions and precision-weighted prediction errors. The network structure remains flexible during message-passing steps, allowing for dynamic adjustments. They can be used as a biologically plausible cognitive model in computational neuroscience or as a generalization of Bayesian filtering for designing efficient, modular decision-making agents. The default implementation supports the generalized Hierarchical Gaussian Filters (gHGF, Weber et al., 2024), but the framework is designed to be adaptable to other algorithms. Built on top of JAX, the core functions are differentiable and JIT-compiled where applicable. The library is optimized for modularity and ease of use, allowing seamless integration with other libraries in the ecosystem for Bayesian inference and optimization. Additionally, a binding with an implementation in Rust is under active development, which will further enhance flexibility during inference. You can find the method paper describing the toolbox [here](https://arxiv.org/abs/2410.09206) and the method paper describing the gHGF, which is the main framework currently supported by the toolbox [here](https://arxiv.org/abs/2305.10937).
-* 📖 [API Documentation](https://ilabcode.github.io/pyhgf/api.html)
-* ✏️ [Tutorials and examples](https://ilabcode.github.io/pyhgf/learn.html)
+* 📖 [API Documentation](https://computationalpsychiatry.github.io/pyhgf/api.html)
+* ✏️ [Tutorials and examples](https://computationalpsychiatryionalpsychiatry.github.io/pyhgf/learn.html)
## Getting started
@@ -21,7 +21,7 @@ The last official release can be downloaded from PIP:
The current version under development can be installed from the master branch of the GitHub folder:
-`pip install “git+https://github.com/ilabcode/pyhgf.git”`
+`pip install “git+https://github.com/ComputationalPsychiatry/pyhgf.git”`
### How does it work?
@@ -32,12 +32,12 @@ Dynamic networks can be defined as a tuple containing the following variables:
* A set of update functions. An update function receive a network tuple and returns an updated network tuple.
* An update sequence (tuple) that defines the order and target of the update functions.
- +
+ You can find a deeper introduction to how to create and manipulate networks under the following link:
-* 🎓 [Creating and manipulating networks of probabilistic nodes](https://ilabcode.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
+* 🎓 [Creating and manipulating networks of probabilistic nodes](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
### The Generalized Hierarchical Gaussian Filter
@@ -46,7 +46,7 @@ Generalized Hierarchical Gaussian Filters (gHGF) are specific instances of dynam
You can find a deeper introduction on how does the gHGF works under the following link:
-* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
+* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
### Model fitting
@@ -73,7 +73,7 @@ hgf.input_data(input_data=u)
hgf.plot_trajectories();
```
-
+
```python
from pyhgf.response import binary_softmax_inverse_temperature
@@ -93,7 +93,7 @@ print(f"Sum of surprises = {surprise.sum()}")
## Acknowledgments
-This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ilabcode/HGF.jl).
+This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ComputationalPsychiatry/HGF.jl).
## References
diff --git a/docs/source/conf.py b/docs/source/conf.py
index 839ef49ab..3dd718eab 100644
--- a/docs/source/conf.py
+++ b/docs/source/conf.py
@@ -95,7 +95,7 @@
"icon_links": [
dict(
name="GitHub",
- url="https://github.com/ilabcode/pyhgf",
+ url="https://github.com/ComputationalPsychiatry/pyhgf",
icon="fa-brands fa-square-github",
),
dict(
diff --git a/docs/source/index.md b/docs/source/index.md
index 6d8b78ef7..95225a926 100644
--- a/docs/source/index.md
+++ b/docs/source/index.md
@@ -1,14 +1,14 @@
-[](https://github.com/pre-commit/pre-commit) [](https://github.com/ilabcode/pyhgf/blob/master/LICENSE) [](https://codecov.io/gh/ilabcode/pyhgf) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/) [](https://badge.fury.io/py/pyhgf)
+[](https://github.com/pre-commit/pre-commit) [](https://github.com/ComputationalPsychiatry/pyhgf/blob/master/LICENSE) [](https://codecov.io/gh/ComputationalPsychiatry/pyhgf) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/) [](https://badge.fury.io/py/pyhgf)
# PyHGF: A Neural Network Library for Predictive Coding
-
You can find a deeper introduction to how to create and manipulate networks under the following link:
-* 🎓 [Creating and manipulating networks of probabilistic nodes](https://ilabcode.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
+* 🎓 [Creating and manipulating networks of probabilistic nodes](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
### The Generalized Hierarchical Gaussian Filter
@@ -46,7 +46,7 @@ Generalized Hierarchical Gaussian Filters (gHGF) are specific instances of dynam
You can find a deeper introduction on how does the gHGF works under the following link:
-* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
+* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
### Model fitting
@@ -73,7 +73,7 @@ hgf.input_data(input_data=u)
hgf.plot_trajectories();
```
-
+
```python
from pyhgf.response import binary_softmax_inverse_temperature
@@ -93,7 +93,7 @@ print(f"Sum of surprises = {surprise.sum()}")
## Acknowledgments
-This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ilabcode/HGF.jl).
+This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ComputationalPsychiatry/HGF.jl).
## References
diff --git a/docs/source/conf.py b/docs/source/conf.py
index 839ef49ab..3dd718eab 100644
--- a/docs/source/conf.py
+++ b/docs/source/conf.py
@@ -95,7 +95,7 @@
"icon_links": [
dict(
name="GitHub",
- url="https://github.com/ilabcode/pyhgf",
+ url="https://github.com/ComputationalPsychiatry/pyhgf",
icon="fa-brands fa-square-github",
),
dict(
diff --git a/docs/source/index.md b/docs/source/index.md
index 6d8b78ef7..95225a926 100644
--- a/docs/source/index.md
+++ b/docs/source/index.md
@@ -1,14 +1,14 @@
-[](https://github.com/pre-commit/pre-commit) [](https://github.com/ilabcode/pyhgf/blob/master/LICENSE) [](https://codecov.io/gh/ilabcode/pyhgf) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/) [](https://badge.fury.io/py/pyhgf)
+[](https://github.com/pre-commit/pre-commit) [](https://github.com/ComputationalPsychiatry/pyhgf/blob/master/LICENSE) [](https://codecov.io/gh/ComputationalPsychiatry/pyhgf) [](https://github.com/psf/black) [](http://mypy-lang.org/) [](https://pycqa.github.io/isort/) [](https://badge.fury.io/py/pyhgf)
# PyHGF: A Neural Network Library for Predictive Coding
- +
+ PyHGF is a Python library for creating and manipulating dynamic probabilistic networks for predictive coding. These networks approximate Bayesian inference by optimizing beliefs through the diffusion of predictions and precision-weighted prediction errors. The network structure remains flexible during message-passing steps, allowing for dynamic adjustments. They can be used as a biologically plausible cognitive model in computational neuroscience or as a generalization of Bayesian filtering for designing efficient, modular decision-making agents. The default implementation supports the generalized Hierarchical Gaussian Filters (gHGF, Weber et al., 2024), but the framework is designed to be adaptable to other algorithms. Built on top of JAX, the core functions are differentiable and JIT-compiled where applicable. The library is optimized for modularity and ease of use, allowing seamless integration with other libraries in the ecosystem for Bayesian inference and optimization. Additionally, a binding with an implementation in Rust is under active development, which will further enhance flexibility during inference. You can find the method paper describing the toolbox [here](https://arxiv.org/abs/2410.09206) and the method paper describing the gHGF, which is the main framework currently supported by the toolbox [here](https://arxiv.org/abs/2305.10937).
-* 📖 [API Documentation](https://ilabcode.github.io/pyhgf/api.html)
-* ✏️ [Tutorials and examples](https://ilabcode.github.io/pyhgf/learn.html)
+* 📖 [API Documentation](https://computationalpsychiatry.github.io/pyhgf/api.html)
+* ✏️ [Tutorials and examples](https://computationalpsychiatry.github.io/pyhgf/learn.html)
## Getting started
@@ -23,7 +23,7 @@ pip install pyhgf
The current version under development can be installed from the master branch of the GitHub folder:
```bash
-pip install “git+https://github.com/ilabcode/pyhgf.git”
+pip install “git+https://github.com/ComputationalPsychiatry/pyhgf.git”
```
### How does it work?
@@ -35,12 +35,12 @@ Dynamic networks can be defined as a tuple containing the following variables:
* A set of update functions. An update function receive a network tuple and returns an updated network tuple.
* An update sequence (tuple) that defines the order and target of the update functions.
-
PyHGF is a Python library for creating and manipulating dynamic probabilistic networks for predictive coding. These networks approximate Bayesian inference by optimizing beliefs through the diffusion of predictions and precision-weighted prediction errors. The network structure remains flexible during message-passing steps, allowing for dynamic adjustments. They can be used as a biologically plausible cognitive model in computational neuroscience or as a generalization of Bayesian filtering for designing efficient, modular decision-making agents. The default implementation supports the generalized Hierarchical Gaussian Filters (gHGF, Weber et al., 2024), but the framework is designed to be adaptable to other algorithms. Built on top of JAX, the core functions are differentiable and JIT-compiled where applicable. The library is optimized for modularity and ease of use, allowing seamless integration with other libraries in the ecosystem for Bayesian inference and optimization. Additionally, a binding with an implementation in Rust is under active development, which will further enhance flexibility during inference. You can find the method paper describing the toolbox [here](https://arxiv.org/abs/2410.09206) and the method paper describing the gHGF, which is the main framework currently supported by the toolbox [here](https://arxiv.org/abs/2305.10937).
-* 📖 [API Documentation](https://ilabcode.github.io/pyhgf/api.html)
-* ✏️ [Tutorials and examples](https://ilabcode.github.io/pyhgf/learn.html)
+* 📖 [API Documentation](https://computationalpsychiatry.github.io/pyhgf/api.html)
+* ✏️ [Tutorials and examples](https://computationalpsychiatry.github.io/pyhgf/learn.html)
## Getting started
@@ -23,7 +23,7 @@ pip install pyhgf
The current version under development can be installed from the master branch of the GitHub folder:
```bash
-pip install “git+https://github.com/ilabcode/pyhgf.git”
+pip install “git+https://github.com/ComputationalPsychiatry/pyhgf.git”
```
### How does it work?
@@ -35,12 +35,12 @@ Dynamic networks can be defined as a tuple containing the following variables:
* A set of update functions. An update function receive a network tuple and returns an updated network tuple.
* An update sequence (tuple) that defines the order and target of the update functions.
- +
+ You can find a deeper introduction to how to create and manipulate networks under the following link:
-* 🎓 [Creating and manipulating networks of probabilistic nodes](https://ilabcode.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
+* 🎓 [Creating and manipulating networks of probabilistic nodes](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
### The Generalized Hierarchical Gaussian Filter
@@ -49,7 +49,7 @@ Generalized Hierarchical Gaussian Filters (gHGF) are specific instances of dynam
You can find a deeper introduction on how does the gHGF works under the following link:
-* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
+* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
### Model fitting
@@ -76,7 +76,7 @@ hgf.input_data(input_data=u)
hgf.plot_trajectories();
```
-
+
```python
from pyhgf.response import binary_softmax_inverse_temperature
@@ -96,7 +96,7 @@ print(f"Sum of surprises = {surprise.sum()}")
## Acknowledgments
-This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ilabcode/HGF.jl).
+This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ComputationalPsychiatry/HGF.jl).
## References
diff --git a/docs/source/learn.md b/docs/source/learn.md
index f99e8672c..076e1f8cb 100644
--- a/docs/source/learn.md
+++ b/docs/source/learn.md
@@ -50,7 +50,7 @@ notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
# Learn
-In this section, you can find tutorial notebooks that describe the internals of pyhgf, the theory behind the Hierarchical Gaussian filter, and step-by-step application and use cases of the model. At the beginning of every tutorial, you will find a badge [](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.1-Creating_networks.ipynb) to run the notebook interactively in a Google Colab session.
+In this section, you can find tutorial notebooks that describe the internals of pyhgf, the theory behind the Hierarchical Gaussian filter, and step-by-step application and use cases of the model. At the beginning of every tutorial, you will find a badge [](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.1-Creating_networks.ipynb) to run the notebook interactively in a Google Colab session.
## Theory
diff --git a/docs/source/notebooks/0.1-Theory.ipynb b/docs/source/notebooks/0.1-Theory.ipynb
index c995adf49..af2327fbe 100644
--- a/docs/source/notebooks/0.1-Theory.ipynb
+++ b/docs/source/notebooks/0.1-Theory.ipynb
@@ -14,7 +14,7 @@
"(theory)=\n",
"# Introduction to the Generalised Hierarchical Gaussian Filter\n",
"\n",
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.1-Theory.ipynb)\n",
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.1-Theory.ipynb)\n",
"\n",
"The Hierarchical Gaussian Filter is a Bayesian model of belief updating under uncertainty in which the volatility of the environment is encoded by a hierarchy of probability distributions. A generalised version of this model {cite:p}`weber:2023` has further framed this into a neural network framework of probabilistic state nodes interacting with each other. At the heart of both frameworks lies a generative model that consist in a hierarchy of {term}`Gaussian Random Walk`. The inversion of this model (i.e. estimating parameters from observed values) leads to the derivation of prediction errors and posterior updates that can turn these networks into learning systems. To fully understand this model, it is therefore central to start with the simplest case, the implied generative model, which can be seen as the probabilistic structure that generates observations.\n",
"\n",
@@ -664,7 +664,7 @@
"source": [
"### The propagation of prediction and prediction errors\n",
"\n",
- "Having described the model as a specific configuration of predictive nodes offer many advantages, especially in term of modularity for the user. However, the model itself is not limited to the description of the generative process that we covered in the previous examples. The most interesting, and also the more complex, part of the modelling consists of the capability for the network to update the hierarchical structure in a Bayesian optimal way as new observations are presented. These steps are defined by a set of simple, one-step update equations that represent changes in beliefs about the hidden states (i.e. the sufficient statistics of the nodes) specified in the generative model. These equations were first described in {cite:t}`2011:mathys`, and the update equations for volatility and value coupling in the generalized Hierarchical Gaussian filter (on which most of the update functions in [pyhgf](https://github.com/ilabcode/pyhgf) are based) have been described in {cite:p}`weber:2023`. The exact computations in each step especially depend on the nature of the coupling (via {term}`VAPE`s vs. {term}`VOPE`s) between the parent and children nodes. It is beyond the scope of this tutorial to dive into the derivation of these steps and we refer the interested reader to the mentioned papers. Here, we provide a general overview of the dynamic of the update sequence that supports belief updating. The computations triggered by any observation at each time point can be ordered in time as shown in the belief update algorithm."
+ "Having described the model as a specific configuration of predictive nodes offer many advantages, especially in term of modularity for the user. However, the model itself is not limited to the description of the generative process that we covered in the previous examples. The most interesting, and also the more complex, part of the modelling consists of the capability for the network to update the hierarchical structure in a Bayesian optimal way as new observations are presented. These steps are defined by a set of simple, one-step update equations that represent changes in beliefs about the hidden states (i.e. the sufficient statistics of the nodes) specified in the generative model. These equations were first described in {cite:t}`2011:mathys`, and the update equations for volatility and value coupling in the generalized Hierarchical Gaussian filter (on which most of the update functions in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) are based) have been described in {cite:p}`weber:2023`. The exact computations in each step especially depend on the nature of the coupling (via {term}`VAPE`s vs. {term}`VOPE`s) between the parent and children nodes. It is beyond the scope of this tutorial to dive into the derivation of these steps and we refer the interested reader to the mentioned papers. Here, we provide a general overview of the dynamic of the update sequence that supports belief updating. The computations triggered by any observation at each time point can be ordered in time as shown in the belief update algorithm."
]
},
{
diff --git a/docs/source/notebooks/0.2-Creating_networks.ipynb b/docs/source/notebooks/0.2-Creating_networks.ipynb
index 82dffc43c..c3640e0e5 100644
--- a/docs/source/notebooks/0.2-Creating_networks.ipynb
+++ b/docs/source/notebooks/0.2-Creating_networks.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.2-Creating_networks.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.2-Creating_networks.ipynb)"
]
},
{
@@ -85,7 +85,7 @@
"id": "57dde3f9-d8b1-437e-8a33-7aea1a1b1e2e",
"metadata": {},
"source": [
- "[pyhgf](https://ilabcode.github.io/pyhgf/index.html#) is designed with inspiration from graph neural network libraries that can support message-passing schemes and perform belief propagation through networks of probabilistic nodes. Here, this principle is applied to predictive processing and focuses on networks that are structured as **rooted trees** and perform variational message passing to update beliefs about the state of the environment, inferred from the observations at the root of the tree. While this library is optimized to implement the standard two-level and three-level HGF {cite:p}`2011:mathys,2014:mathys`, as well as the generalized HGF {cite:p}`weber:2023`, it can also be applied to much larger use cases, with the idea is to generalize belief propagation as it has been described so far to larger and more complex networks that will capture a greater variety of environmental structure. Therefore, the library is also designed to facilitate the creation and manipulation of such probabilistic networks. Importantly, here we consider that a probabilistic network should be defined by the following four variables:\n",
+ "[pyhgf](https://computationalpsychiatry.github.io/pyhgf/index.html#) is designed with inspiration from graph neural network libraries that can support message-passing schemes and perform belief propagation through networks of probabilistic nodes. Here, this principle is applied to predictive processing and focuses on networks that are structured as **rooted trees** and perform variational message passing to update beliefs about the state of the environment, inferred from the observations at the root of the tree. While this library is optimized to implement the standard two-level and three-level HGF {cite:p}`2011:mathys,2014:mathys`, as well as the generalized HGF {cite:p}`weber:2023`, it can also be applied to much larger use cases, with the idea is to generalize belief propagation as it has been described so far to larger and more complex networks that will capture a greater variety of environmental structure. Therefore, the library is also designed to facilitate the creation and manipulation of such probabilistic networks. Importantly, here we consider that a probabilistic network should be defined by the following four variables:\n",
"1. the network parameters\n",
"2. the network structure\n",
"3. the update function(s)\n",
@@ -711,7 +711,7 @@
"source": [
"### Multivariate coupling\n",
"\n",
- "As we can see in the examples above, nodes in a valid HGF network can be influenced by multiple parents (either value or volatility parents). Similarly, a single node can be influenced by multiple children. This feature is termed *multivariate descendency* and *multivariate ascendency* (respectively) and is a central addition to the generalization of the HGF {cite:p}`weber:2023` that was implemented in this package, as well as in the [Julia counterpart](https://github.com/ilabcode/HierarchicalGaussianFiltering.jl)."
+ "As we can see in the examples above, nodes in a valid HGF network can be influenced by multiple parents (either value or volatility parents). Similarly, a single node can be influenced by multiple children. This feature is termed *multivariate descendency* and *multivariate ascendency* (respectively) and is a central addition to the generalization of the HGF {cite:p}`weber:2023` that was implemented in this package, as well as in the [Julia counterpart](https://github.com/ComputationalPsychiatry/HierarchicalGaussianFiltering.jl)."
]
},
{
@@ -1748,7 +1748,7 @@
"source": [
"## Creating custom update functions\n",
"\n",
- "The structure of the network and the node's parameters are the most static component of the network. Actually, we could consider that the network already exists once those two variables are in place. However in [pyhgf](https://ilabcode.github.io/pyhgf/index.html#) we consider that the update functions $\\mathcal{F} = \\{f_1, ..., f_n\\}$ and the update sequence $\\Sigma = [f_1(n_1), ..., f_i(n_j), f \\in \\mathcal{F}, n \\in 1, ..., k ]$ (the order of update) are also part of the models. This choice was made to explicitly account that there is no one unique way of modelling the way beliefs propagate through the network, and a core task for predictive coding applications is to develop new *probabilistic nodes* that account for a greater variety of phenomena. This step critically requires modelling beliefs diffusion, and therefore to modify, or creating the underlying update functions."
+ "The structure of the network and the node's parameters are the most static component of the network. Actually, we could consider that the network already exists once those two variables are in place. However in [pyhgf](https://computationalpsychiatry.github.io/pyhgf/index.html#) we consider that the update functions $\\mathcal{F} = \\{f_1, ..., f_n\\}$ and the update sequence $\\Sigma = [f_1(n_1), ..., f_i(n_j), f \\in \\mathcal{F}, n \\in 1, ..., k ]$ (the order of update) are also part of the models. This choice was made to explicitly account that there is no one unique way of modelling the way beliefs propagate through the network, and a core task for predictive coding applications is to develop new *probabilistic nodes* that account for a greater variety of phenomena. This step critically requires modelling beliefs diffusion, and therefore to modify, or creating the underlying update functions."
]
},
{
diff --git a/docs/source/notebooks/0.3-Generalised_filtering.ipynb b/docs/source/notebooks/0.3-Generalised_filtering.ipynb
index 552989459..5452eb9ce 100644
--- a/docs/source/notebooks/0.3-Generalised_filtering.ipynb
+++ b/docs/source/notebooks/0.3-Generalised_filtering.ipynb
@@ -14,7 +14,7 @@
"(generalised_filtering)=\n",
"# From Reinforcement Learning to Generalised Bayesian Filtering\n",
"\n",
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.3-Generalised_filtering.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.3-Generalised_filtering.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/1.1-Binary_HGF.ipynb b/docs/source/notebooks/1.1-Binary_HGF.ipynb
index 83297bccc..5d02d4deb 100644
--- a/docs/source/notebooks/1.1-Binary_HGF.ipynb
+++ b/docs/source/notebooks/1.1-Binary_HGF.ipynb
@@ -20,7 +20,7 @@
"id": "1258ea53-11fc-4a27-927c-2afc91f4aa12",
"metadata": {},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.1-Binary_HGF.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.1-Binary_HGF.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/1.2-Categorical_HGF.ipynb b/docs/source/notebooks/1.2-Categorical_HGF.ipynb
index f4a42850f..06ab288d2 100644
--- a/docs/source/notebooks/1.2-Categorical_HGF.ipynb
+++ b/docs/source/notebooks/1.2-Categorical_HGF.ipynb
@@ -20,7 +20,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.3-CAtegorical_HGF.ipynb)\n",
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.3-CAtegorical_HGF.ipynb)\n",
"\n",
"```{warning}\n",
"The categorical state node and the categorical state-transition nodes are still work in progress. The examples provided here are given for illustration. Things may change or not work until the official publication.\n",
diff --git a/docs/source/notebooks/1.3-Continuous_HGF.ipynb b/docs/source/notebooks/1.3-Continuous_HGF.ipynb
index 84e0def7e..db1e94681 100644
--- a/docs/source/notebooks/1.3-Continuous_HGF.ipynb
+++ b/docs/source/notebooks/1.3-Continuous_HGF.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.3-Continuous_HGF.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.3-Continuous_HGF.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/2-Using_custom_response_functions.ipynb b/docs/source/notebooks/2-Using_custom_response_functions.ipynb
index 71554bcb4..774e43dad 100644
--- a/docs/source/notebooks/2-Using_custom_response_functions.ipynb
+++ b/docs/source/notebooks/2-Using_custom_response_functions.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/2-Using_custom_response_functions.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/2-Using_custom_response_functions.ipynb)"
]
},
{
@@ -542,12 +542,12 @@
"\n",
"Let's now consider that the two vectors of observations $u$ and responses $y$ were obtained from a real participant undergoing a real experiment. In this situation, we assume that this participant internally used a {term}`Perceptual model` and a {term}`Decision rule` that might resemble what we defined previously, and we want to infer what are the most likely values for critical parameters in the model (e.g. the evolution rate $\\omega_2$). To do so, we are going to use our dataset (both $u$ and $y$), and try many models. We are going to fix the values of all HGF parameters to reasonable estimates (here, using the exact same values as in the simulation), except for $\\omega_2$. For this last parameter, we will assume a prior set at $\\mathcal{N}(-2.0, 2.0)$. The idea is that we want to sample many $\\omega_2$ values from this distribution and see if the model is performing better with some values.\n",
"\n",
- "But here, we need a clear definition of what this means *to perform better* for a given model. And this is exactly what a {term}`Response model` does, it is a way for us to evaluate how likely the behaviours $y$ for a given {term}`Perceptual model`, and assuming that the participants use this specific {term}`Decision rule`. In [pyhgf](https://github.com/ilabcode/pyhgf), this step is performed by creating the corresponding {term}`Response function`, which is the Python function that will return the surprise $S$ of getting these behaviours from the participant under this decision rule.\n",
+ "But here, we need a clear definition of what this means *to perform better* for a given model. And this is exactly what a {term}`Response model` does, it is a way for us to evaluate how likely the behaviours $y$ for a given {term}`Perceptual model`, and assuming that the participants use this specific {term}`Decision rule`. In [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), this step is performed by creating the corresponding {term}`Response function`, which is the Python function that will return the surprise $S$ of getting these behaviours from the participant under this decision rule.\n",
"\n",
"````{hint} What is a *response function*?\n",
"Most of the work around HGFs consists in creating and adapting {term}`Response model` to work with a given experimental design. There is no limit in terms of what can be used as a {term}`Response model`, provided that the {term}`Perceptual model` and the {term}`Decision rule` are clearly defined.\n",
"\n",
- "In [pyhgf](https://github.com/ilabcode/pyhgf), the {term}`Perceptual model` is the probabilistic network created with the main {py:class}`pyhgf.model.HGF` and {py:class}`pyhgf.distribution.HGFDistribution` classes. The {term}`Response model` is something that is implicitly defined when we create the {term}`Response function`, a Python function that computes the negative of the log-likelihood of the actions given the perceptual model. This {term}`Response function` can be passed as an argument to the main classes using the keywords arguments `response_function`, `response_function_inputs` and `response_function_parameters`. The `response_function` can be any callable that returns the surprise $S$ of observing action $y$ given this model, and the {term}`Decision rule`. The `response_function_inputs` are the additional data to the response function (optional) while `response_function_parameters` are the additional parameters (optional). The `response_function_inputs` is where the actions $y$ should be provided.\n",
+ "In [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), the {term}`Perceptual model` is the probabilistic network created with the main {py:class}`pyhgf.model.HGF` and {py:class}`pyhgf.distribution.HGFDistribution` classes. The {term}`Response model` is something that is implicitly defined when we create the {term}`Response function`, a Python function that computes the negative of the log-likelihood of the actions given the perceptual model. This {term}`Response function` can be passed as an argument to the main classes using the keywords arguments `response_function`, `response_function_inputs` and `response_function_parameters`. The `response_function` can be any callable that returns the surprise $S$ of observing action $y$ given this model, and the {term}`Decision rule`. The `response_function_inputs` are the additional data to the response function (optional) while `response_function_parameters` are the additional parameters (optional). The `response_function_inputs` is where the actions $y$ should be provided.\n",
"\n",
"```{important}\n",
"A *response function* should not return the actions given perceptual inputs $y | u$ (this is what the {term}`Decision rule` does), but the [surprise](https://en.wikipedia.org/wiki/Information_content) $S$ associated with the observation of actions given the perceptual inputs $S(y | u)$, which is defined by:\n",
@@ -559,7 +559,7 @@
"$$\n",
"```\n",
"\n",
- "If you are already familiar with using HGFs in the Julia equivalent of [pyhgf](https://github.com/ilabcode/pyhgf), you probably noted that the toolbox is split into a **perceptual** package [HierarchicalGaussianFiltering.jl](https://github.com/ilabcode/HierarchicalGaussianFiltering.jl) and a **response** package [ActionModels.jl](https://github.com/ilabcode/ActionModels.jl). This was made to make the difference between the two parts of the HGF clear and be explicit that you can use a perceptual model without any action model. In [pyhgf](https://github.com/ilabcode/pyhgf) however, everything happens in the same package, the response function is merely an optional, additional argument that can be passed to describe how surprise is computed.\n",
+ "If you are already familiar with using HGFs in the Julia equivalent of [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), you probably noted that the toolbox is split into a **perceptual** package [HierarchicalGaussianFiltering.jl](https://github.com/ComputationalPsychiatry/HierarchicalGaussianFiltering.jl) and a **response** package [ActionModels.jl](https://github.com/ComputationalPsychiatry/ActionModels.jl). This was made to make the difference between the two parts of the HGF clear and be explicit that you can use a perceptual model without any action model. In [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) however, everything happens in the same package, the response function is merely an optional, additional argument that can be passed to describe how surprise is computed.\n",
"````"
]
},
@@ -637,7 +637,7 @@
},
"source": [
"```{note}\n",
- "Note here that our {term}`Response function` has a structure that is the standard way to write response functions in [pyhgf](https://github.com/ilabcode/pyhgf), that is with two input arguments:\n",
+ "Note here that our {term}`Response function` has a structure that is the standard way to write response functions in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), that is with two input arguments:\n",
"- the HGF model on which the response function applies (i.e. the {term}`Perceptual model`)\n",
"- the additional parameters provided to the response function. This can include additional parameters that can be part of the equation of the model, or the input data used by this model. We then provide the `response` vector ($y$) here.\n",
"\n",
@@ -1014,7 +1014,7 @@
"```{glossary}\n",
"\n",
"Perceptual model\n",
- " The perceptual model of a Hierarchical Gaussian Filter traditionally refers to the branch receiving observations $u$ about states of the world and that performs the updating of beliefs about these states. By generalisation, the perceptual model is any probabilistic network that can be created in [pyhgf](https://github.com/ilabcode/pyhgf), receiving an arbitrary number of inputs. An HGF that only consists of a perceptual model will act as a Bayesian filter.\n",
+ " The perceptual model of a Hierarchical Gaussian Filter traditionally refers to the branch receiving observations $u$ about states of the world and that performs the updating of beliefs about these states. By generalisation, the perceptual model is any probabilistic network that can be created in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), receiving an arbitrary number of inputs. An HGF that only consists of a perceptual model will act as a Bayesian filter.\n",
"\n",
"Response model\n",
" The response model of a Hierarchical Gaussian filter refers to the branch that uses the beliefs about the state of the world to generate actions using the {term}`Decision rule`. This branch is also sometimes referred to as the **decision model** or the **observation model**, depending on the fields. Critically, this part of the model can return the surprise ($-\\log[Pr(x)]$) associated with the observations (here, the observations include the inputs $u$ of the probabilistic network, but will also include the responses of the participant $y$ if there are some).\n",
@@ -1023,7 +1023,7 @@
" The decision rule is a function stating how the agent selects among all possible actions, given the state of the beliefs in the perceptual model, and optionally additional parameters. Programmatically, this is a Python function taking a perceptual model as input (i.e. an instance of the HGF class), and returning a sequence of actions. This can be used for simulation. The decision rule should be clearly defined in order to write the {term}`Response function`.\n",
"\n",
"Response function\n",
- " The response function is a term that we use specifically for this package ([pyhgf](https://github.com/ilabcode/pyhgf)). It refers to the Python function that, using a given HGF model and optional parameter, returns the surprise associated with the observed actions.\n",
+ " The response function is a term that we use specifically for this package ([pyhgf](https://github.com/ComputationalPsychiatry/pyhgf)). It refers to the Python function that, using a given HGF model and optional parameter, returns the surprise associated with the observed actions.\n",
"\n",
"```"
]
diff --git a/docs/source/notebooks/3-Multilevel_HGF.ipynb b/docs/source/notebooks/3-Multilevel_HGF.ipynb
index 691aea4eb..c7abd2603 100644
--- a/docs/source/notebooks/3-Multilevel_HGF.ipynb
+++ b/docs/source/notebooks/3-Multilevel_HGF.ipynb
@@ -32,7 +32,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/3-Multilevel_HGF.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/3-Multilevel_HGF.ipynb)"
]
},
{
@@ -137,7 +137,7 @@
"Luckily, we already have all the components in place to do that. We already used Bayesian networks in the previous sections when we were inferring the distribution of some parameters. Here, we only had one agent (i.e. one participant), and therefore did not need any hyperprior. We need to extend this approach a bit, and explicitly state that we want to fit many models (participants) simultaneously, and draw the values of some parameters from a hyper-prior (i.e. the group-level distribution).\n",
"\n",
"But before we move forward, maybe it is worth clarifying some of the terminology we use, especially as, starting from now, many things are called **networks** but are pointing to different parts of the workflow. We can indeed distinguish two kinds:\n",
- "1. The predictive coding neural networks. This is the kind of network that [pyhgf](https://github.com/ilabcode/pyhgf) is designed to handle (see {ref}`probabilistic_networks`). Every HGF model is an instance of such a network.\n",
+ "1. The predictive coding neural networks. This is the kind of network that [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) is designed to handle (see {ref}`probabilistic_networks`). Every HGF model is an instance of such a network.\n",
"2. The Bayesian (multilevel) network is the computational graph that is created with tools like [pymc](https://www.pymc.io/welcome.html). This graph will represent the dependencies between our variables and the way they are transformed.\n",
"\n",
"In this notebook, we are going to create the second type of network and incorporate many networks of the first type in it as custom distribution."
diff --git a/docs/source/notebooks/4-Parameter_recovery.ipynb b/docs/source/notebooks/4-Parameter_recovery.ipynb
index 1120471f0..f019c30d4 100644
--- a/docs/source/notebooks/4-Parameter_recovery.ipynb
+++ b/docs/source/notebooks/4-Parameter_recovery.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/4-Parameter_recovery.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/4-Parameter_recovery.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/5-Non_linear_value_coupling.ipynb b/docs/source/notebooks/5-Non_linear_value_coupling.ipynb
index 1daa5cdf7..cb78a2d8c 100644
--- a/docs/source/notebooks/5-Non_linear_value_coupling.ipynb
+++ b/docs/source/notebooks/5-Non_linear_value_coupling.ipynb
@@ -12,7 +12,7 @@
"cell_type": "markdown",
"metadata": {},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.4-Continuous_HGF_non_linear_value_coupling.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.4-Continuous_HGF_non_linear_value_coupling.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb b/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb
index 8ef988a6d..8078c3a78 100644
--- a/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb
+++ b/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb)"
]
},
{
@@ -106,7 +106,7 @@
"id": "48020213-7f26-4201-b4b9-f072231bc225",
"metadata": {},
"source": [
- "The nodalized version of the Hierarchical Gaussian Filter that is implemented in [pyhgf](https://github.com/ilabcode/pyhgf) opens the possibility to create filters with multiple inputs. Here, we illustrate how we can use this feature to create an agent that is filtering their physiological signals in real-time. We use a two-level Hierarchical Gaussian Filter to predict the dynamics of the instantaneous heart rate (the RR interval measured at each heartbeat). We then extract the trajectory of surprise at each predictive node to relate it with the cognitive task performed by the participant while the signal is being recorded."
+ "The nodalized version of the Hierarchical Gaussian Filter that is implemented in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) opens the possibility to create filters with multiple inputs. Here, we illustrate how we can use this feature to create an agent that is filtering their physiological signals in real-time. We use a two-level Hierarchical Gaussian Filter to predict the dynamics of the instantaneous heart rate (the RR interval measured at each heartbeat). We then extract the trajectory of surprise at each predictive node to relate it with the cognitive task performed by the participant while the signal is being recorded."
]
},
{
diff --git a/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb b/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb
index b11635c5a..5bbf09532 100644
--- a/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb
+++ b/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb b/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb
index 4ce529bfa..955bde00a 100644
--- a/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb
+++ b/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb b/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb
index 03c230d81..bc126d259 100644
--- a/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb
+++ b/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb)"
]
},
{
@@ -137,7 +137,7 @@
"x_1^{(k)} \\sim \\mathcal{N}(x_1^{(k-1)}, \\sigma^2)\n",
"$$\n",
"\n",
- "where $\\sigma^2$ is the fixed variance of the distribution. You can find more details on this as well as some code to get started with the simulations in the first tutorial on the [PyHGF documentation](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#).\n",
+ "where $\\sigma^2$ is the fixed variance of the distribution. You can find more details on this as well as some code to get started with the simulations in the first tutorial on the [PyHGF documentation](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#).\n",
"\n",
"```{exercise}\n",
":label: exercise1.1\n",
@@ -183,7 +183,7 @@
"$$\n",
"\n",
"```{hint}\n",
- "This generative process reads top-down: the node higher in the hierarchy ($x_2$) generates new values and passes them to the child nodes. We can also control how much the node $x_1$ is influenced by its previous (see examples [here](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#value-coupling)). To keep things simple in this exercise, we will assume that $x_1$ is fully influenced by its previous value, as noted in the equation, therefore $x_2$ can be seen as a *drift parent*.\n",
+ "This generative process reads top-down: the node higher in the hierarchy ($x_2$) generates new values and passes them to the child nodes. We can also control how much the node $x_1$ is influenced by its previous (see examples [here](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#value-coupling)). To keep things simple in this exercise, we will assume that $x_1$ is fully influenced by its previous value, as noted in the equation, therefore $x_2$ can be seen as a *drift parent*.\n",
"```\n",
"\n",
"Let's define a time series `x_2` that represent the evolution of the states of the parent node:"
@@ -373,11 +373,11 @@
"source": [
"Building on these principles, we can create networks of arbitrary size and shape made of probabilistic nodes (nodes that represent the evolution of a Gaussian Random Walk) that can be connected with each other through **value** or **volatility** coupling. We have covered these points in the previous exercise using a simplified network made of two nodes.\n",
"\n",
- "[PyHGF](https://ilabcode.github.io/pyhgf/index.html) adds most of its values in that it allows the creation and manipulation of large and complex networks easily, and it automates the transmission of values between nodes. You can refer to [the second part of the tutorial documentation](https://ilabcode.github.io/pyhgf/notebooks/0.2-Creating_networks.html) which covers the manipulation of networks for more details. \n",
+ "[PyHGF](https://computationalpsychiatry.github.io/pyhgf/index.html) adds most of its values in that it allows the creation and manipulation of large and complex networks easily, and it automates the transmission of values between nodes. You can refer to [the second part of the tutorial documentation](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.2-Creating_networks.html) which covers the manipulation of networks for more details. \n",
"\n",
"But this is not the only thing it does. So far we have simulated observations from the generative model, going for the leaves to the root of the network. This describes how we expect the environment to behave, but not how an agent would learn from it. To do so we need to invert this model: we want to provide the observations, and let the network update the nodes accordingly, so the beliefs always reflect the current state of the environment. This process requires propagating precision prediction errors from the root to the leaves of the network, and this is where most of PyHGF's dark magic is most useful.\n",
"\n",
- "```{figure} https://raw.githubusercontent.com/ilabcode/pyhgf/master/docs/source/images/graph_network.svg\n",
+ "```{figure} https://raw.githubusercontent.com/ComputationalPsychiatry/pyhgf/master/docs/source/images/graph_network.svg\n",
"---\n",
"name: networks\n",
"---\n",
@@ -760,7 +760,7 @@
],
"source": [
"aarhus_weather_df = pd.read_csv(\n",
- " \"https://raw.githubusercontent.com/ilabcode/hgf-data/main/datasets/weather.csv\"\n",
+ " \"https://raw.githubusercontent.com/ComputationalPsychiatry/hgf-data/main/datasets/weather.csv\"\n",
")\n",
"aarhus_weather_df.head()"
]
@@ -1076,7 +1076,7 @@
"````{solution} exercise1.5\n",
":label: solution-exercise1.5\n",
"\n",
- "This method return the Gaussian surprise at each time point, which are then summed. The sum of the Gaussian surprise reflect the performances of the model at predicting the next value, larger values pointing to a model more surprise by its inputs, therefore poor performances. The surprise is define as $s = -log(p)$, this is thus the negative of the log probability function. Log probability functions are commonly used by sampling algorithm, it is thus straigthforwars to sample a model parameters when this function is available. There are an infinity of response possible functions - just like there is an infinity of possible networks - for more details on how to use tem, you can refer to the [tutorial on custom response models](https://ilabcode.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html).\n",
+ "This method return the Gaussian surprise at each time point, which are then summed. The sum of the Gaussian surprise reflect the performances of the model at predicting the next value, larger values pointing to a model more surprise by its inputs, therefore poor performances. The surprise is define as $s = -log(p)$, this is thus the negative of the log probability function. Log probability functions are commonly used by sampling algorithm, it is thus straigthforwars to sample a model parameters when this function is available. There are an infinity of response possible functions - just like there is an infinity of possible networks - for more details on how to use tem, you can refer to the [tutorial on custom response models](https://computationalpsychiatry.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html).\n",
"\n",
"````"
]
diff --git a/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb b/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
index 7a1b0f4c1..0dc3ce8c6 100644
--- a/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
+++ b/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb)"
]
},
{
@@ -1942,9 +1942,9 @@
"\n",
"All these sections should give you a solid understanding of the model and how to use it in context. If you want to apply this to your dataset, we recommend exploring the tutorial section of the documentation:\n",
"\n",
- "- [Using custom response models](https://ilabcode.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html) will explore the creation of custom models for behaviours that can match your experimental design and theory.\n",
- "- [Multilevel modelling](https://ilabcode.github.io/pyhgf/notebooks/3-Multilevel_HGF.html) will discuss modelling at the group/condition level in multilevel Bayesian networks.\n",
- "- [Parameter recovery](https://ilabcode.github.io/pyhgf/notebooks/4-Parameter_recovery.html) explains how to simulate a dataset and perform parameter recovery, as a prior validation of your models."
+ "- [Using custom response models](https://computationalpsychiatry.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html) will explore the creation of custom models for behaviours that can match your experimental design and theory.\n",
+ "- [Multilevel modelling](https://computationalpsychiatry.github.io/pyhgf/notebooks/3-Multilevel_HGF.html) will discuss modelling at the group/condition level in multilevel Bayesian networks.\n",
+ "- [Parameter recovery](https://computationalpsychiatry.github.io/pyhgf/notebooks/4-Parameter_recovery.html) explains how to simulate a dataset and perform parameter recovery, as a prior validation of your models."
]
},
{
diff --git a/pyproject.toml b/pyproject.toml
index 678d407fd..d9a4246d3 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -4,8 +4,8 @@ description = "Dynamic neural networks for predictive coding"
authors = ["Nicolas Legrand
You can find a deeper introduction to how to create and manipulate networks under the following link:
-* 🎓 [Creating and manipulating networks of probabilistic nodes](https://ilabcode.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
+* 🎓 [Creating and manipulating networks of probabilistic nodes](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.2-Creating_networks.html)
### The Generalized Hierarchical Gaussian Filter
@@ -49,7 +49,7 @@ Generalized Hierarchical Gaussian Filters (gHGF) are specific instances of dynam
You can find a deeper introduction on how does the gHGF works under the following link:
-* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
+* 🎓 [Introduction to the Hierarchical Gaussian Filter](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#theory)
### Model fitting
@@ -76,7 +76,7 @@ hgf.input_data(input_data=u)
hgf.plot_trajectories();
```
-
+
```python
from pyhgf.response import binary_softmax_inverse_temperature
@@ -96,7 +96,7 @@ print(f"Sum of surprises = {surprise.sum()}")
## Acknowledgments
-This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ilabcode/HGF.jl).
+This implementation of the Hierarchical Gaussian Filter was inspired by the original [Matlab HGF Toolbox](https://translationalneuromodeling.github.io/tapas). A Julia implementation is also available [here](https://github.com/ComputationalPsychiatry/HGF.jl).
## References
diff --git a/docs/source/learn.md b/docs/source/learn.md
index f99e8672c..076e1f8cb 100644
--- a/docs/source/learn.md
+++ b/docs/source/learn.md
@@ -50,7 +50,7 @@ notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
# Learn
-In this section, you can find tutorial notebooks that describe the internals of pyhgf, the theory behind the Hierarchical Gaussian filter, and step-by-step application and use cases of the model. At the beginning of every tutorial, you will find a badge [](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.1-Creating_networks.ipynb) to run the notebook interactively in a Google Colab session.
+In this section, you can find tutorial notebooks that describe the internals of pyhgf, the theory behind the Hierarchical Gaussian filter, and step-by-step application and use cases of the model. At the beginning of every tutorial, you will find a badge [](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.1-Creating_networks.ipynb) to run the notebook interactively in a Google Colab session.
## Theory
diff --git a/docs/source/notebooks/0.1-Theory.ipynb b/docs/source/notebooks/0.1-Theory.ipynb
index c995adf49..af2327fbe 100644
--- a/docs/source/notebooks/0.1-Theory.ipynb
+++ b/docs/source/notebooks/0.1-Theory.ipynb
@@ -14,7 +14,7 @@
"(theory)=\n",
"# Introduction to the Generalised Hierarchical Gaussian Filter\n",
"\n",
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.1-Theory.ipynb)\n",
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.1-Theory.ipynb)\n",
"\n",
"The Hierarchical Gaussian Filter is a Bayesian model of belief updating under uncertainty in which the volatility of the environment is encoded by a hierarchy of probability distributions. A generalised version of this model {cite:p}`weber:2023` has further framed this into a neural network framework of probabilistic state nodes interacting with each other. At the heart of both frameworks lies a generative model that consist in a hierarchy of {term}`Gaussian Random Walk`. The inversion of this model (i.e. estimating parameters from observed values) leads to the derivation of prediction errors and posterior updates that can turn these networks into learning systems. To fully understand this model, it is therefore central to start with the simplest case, the implied generative model, which can be seen as the probabilistic structure that generates observations.\n",
"\n",
@@ -664,7 +664,7 @@
"source": [
"### The propagation of prediction and prediction errors\n",
"\n",
- "Having described the model as a specific configuration of predictive nodes offer many advantages, especially in term of modularity for the user. However, the model itself is not limited to the description of the generative process that we covered in the previous examples. The most interesting, and also the more complex, part of the modelling consists of the capability for the network to update the hierarchical structure in a Bayesian optimal way as new observations are presented. These steps are defined by a set of simple, one-step update equations that represent changes in beliefs about the hidden states (i.e. the sufficient statistics of the nodes) specified in the generative model. These equations were first described in {cite:t}`2011:mathys`, and the update equations for volatility and value coupling in the generalized Hierarchical Gaussian filter (on which most of the update functions in [pyhgf](https://github.com/ilabcode/pyhgf) are based) have been described in {cite:p}`weber:2023`. The exact computations in each step especially depend on the nature of the coupling (via {term}`VAPE`s vs. {term}`VOPE`s) between the parent and children nodes. It is beyond the scope of this tutorial to dive into the derivation of these steps and we refer the interested reader to the mentioned papers. Here, we provide a general overview of the dynamic of the update sequence that supports belief updating. The computations triggered by any observation at each time point can be ordered in time as shown in the belief update algorithm."
+ "Having described the model as a specific configuration of predictive nodes offer many advantages, especially in term of modularity for the user. However, the model itself is not limited to the description of the generative process that we covered in the previous examples. The most interesting, and also the more complex, part of the modelling consists of the capability for the network to update the hierarchical structure in a Bayesian optimal way as new observations are presented. These steps are defined by a set of simple, one-step update equations that represent changes in beliefs about the hidden states (i.e. the sufficient statistics of the nodes) specified in the generative model. These equations were first described in {cite:t}`2011:mathys`, and the update equations for volatility and value coupling in the generalized Hierarchical Gaussian filter (on which most of the update functions in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) are based) have been described in {cite:p}`weber:2023`. The exact computations in each step especially depend on the nature of the coupling (via {term}`VAPE`s vs. {term}`VOPE`s) between the parent and children nodes. It is beyond the scope of this tutorial to dive into the derivation of these steps and we refer the interested reader to the mentioned papers. Here, we provide a general overview of the dynamic of the update sequence that supports belief updating. The computations triggered by any observation at each time point can be ordered in time as shown in the belief update algorithm."
]
},
{
diff --git a/docs/source/notebooks/0.2-Creating_networks.ipynb b/docs/source/notebooks/0.2-Creating_networks.ipynb
index 82dffc43c..c3640e0e5 100644
--- a/docs/source/notebooks/0.2-Creating_networks.ipynb
+++ b/docs/source/notebooks/0.2-Creating_networks.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.2-Creating_networks.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.2-Creating_networks.ipynb)"
]
},
{
@@ -85,7 +85,7 @@
"id": "57dde3f9-d8b1-437e-8a33-7aea1a1b1e2e",
"metadata": {},
"source": [
- "[pyhgf](https://ilabcode.github.io/pyhgf/index.html#) is designed with inspiration from graph neural network libraries that can support message-passing schemes and perform belief propagation through networks of probabilistic nodes. Here, this principle is applied to predictive processing and focuses on networks that are structured as **rooted trees** and perform variational message passing to update beliefs about the state of the environment, inferred from the observations at the root of the tree. While this library is optimized to implement the standard two-level and three-level HGF {cite:p}`2011:mathys,2014:mathys`, as well as the generalized HGF {cite:p}`weber:2023`, it can also be applied to much larger use cases, with the idea is to generalize belief propagation as it has been described so far to larger and more complex networks that will capture a greater variety of environmental structure. Therefore, the library is also designed to facilitate the creation and manipulation of such probabilistic networks. Importantly, here we consider that a probabilistic network should be defined by the following four variables:\n",
+ "[pyhgf](https://computationalpsychiatry.github.io/pyhgf/index.html#) is designed with inspiration from graph neural network libraries that can support message-passing schemes and perform belief propagation through networks of probabilistic nodes. Here, this principle is applied to predictive processing and focuses on networks that are structured as **rooted trees** and perform variational message passing to update beliefs about the state of the environment, inferred from the observations at the root of the tree. While this library is optimized to implement the standard two-level and three-level HGF {cite:p}`2011:mathys,2014:mathys`, as well as the generalized HGF {cite:p}`weber:2023`, it can also be applied to much larger use cases, with the idea is to generalize belief propagation as it has been described so far to larger and more complex networks that will capture a greater variety of environmental structure. Therefore, the library is also designed to facilitate the creation and manipulation of such probabilistic networks. Importantly, here we consider that a probabilistic network should be defined by the following four variables:\n",
"1. the network parameters\n",
"2. the network structure\n",
"3. the update function(s)\n",
@@ -711,7 +711,7 @@
"source": [
"### Multivariate coupling\n",
"\n",
- "As we can see in the examples above, nodes in a valid HGF network can be influenced by multiple parents (either value or volatility parents). Similarly, a single node can be influenced by multiple children. This feature is termed *multivariate descendency* and *multivariate ascendency* (respectively) and is a central addition to the generalization of the HGF {cite:p}`weber:2023` that was implemented in this package, as well as in the [Julia counterpart](https://github.com/ilabcode/HierarchicalGaussianFiltering.jl)."
+ "As we can see in the examples above, nodes in a valid HGF network can be influenced by multiple parents (either value or volatility parents). Similarly, a single node can be influenced by multiple children. This feature is termed *multivariate descendency* and *multivariate ascendency* (respectively) and is a central addition to the generalization of the HGF {cite:p}`weber:2023` that was implemented in this package, as well as in the [Julia counterpart](https://github.com/ComputationalPsychiatry/HierarchicalGaussianFiltering.jl)."
]
},
{
@@ -1748,7 +1748,7 @@
"source": [
"## Creating custom update functions\n",
"\n",
- "The structure of the network and the node's parameters are the most static component of the network. Actually, we could consider that the network already exists once those two variables are in place. However in [pyhgf](https://ilabcode.github.io/pyhgf/index.html#) we consider that the update functions $\\mathcal{F} = \\{f_1, ..., f_n\\}$ and the update sequence $\\Sigma = [f_1(n_1), ..., f_i(n_j), f \\in \\mathcal{F}, n \\in 1, ..., k ]$ (the order of update) are also part of the models. This choice was made to explicitly account that there is no one unique way of modelling the way beliefs propagate through the network, and a core task for predictive coding applications is to develop new *probabilistic nodes* that account for a greater variety of phenomena. This step critically requires modelling beliefs diffusion, and therefore to modify, or creating the underlying update functions."
+ "The structure of the network and the node's parameters are the most static component of the network. Actually, we could consider that the network already exists once those two variables are in place. However in [pyhgf](https://computationalpsychiatry.github.io/pyhgf/index.html#) we consider that the update functions $\\mathcal{F} = \\{f_1, ..., f_n\\}$ and the update sequence $\\Sigma = [f_1(n_1), ..., f_i(n_j), f \\in \\mathcal{F}, n \\in 1, ..., k ]$ (the order of update) are also part of the models. This choice was made to explicitly account that there is no one unique way of modelling the way beliefs propagate through the network, and a core task for predictive coding applications is to develop new *probabilistic nodes* that account for a greater variety of phenomena. This step critically requires modelling beliefs diffusion, and therefore to modify, or creating the underlying update functions."
]
},
{
diff --git a/docs/source/notebooks/0.3-Generalised_filtering.ipynb b/docs/source/notebooks/0.3-Generalised_filtering.ipynb
index 552989459..5452eb9ce 100644
--- a/docs/source/notebooks/0.3-Generalised_filtering.ipynb
+++ b/docs/source/notebooks/0.3-Generalised_filtering.ipynb
@@ -14,7 +14,7 @@
"(generalised_filtering)=\n",
"# From Reinforcement Learning to Generalised Bayesian Filtering\n",
"\n",
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/0.3-Generalised_filtering.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/0.3-Generalised_filtering.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/1.1-Binary_HGF.ipynb b/docs/source/notebooks/1.1-Binary_HGF.ipynb
index 83297bccc..5d02d4deb 100644
--- a/docs/source/notebooks/1.1-Binary_HGF.ipynb
+++ b/docs/source/notebooks/1.1-Binary_HGF.ipynb
@@ -20,7 +20,7 @@
"id": "1258ea53-11fc-4a27-927c-2afc91f4aa12",
"metadata": {},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.1-Binary_HGF.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.1-Binary_HGF.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/1.2-Categorical_HGF.ipynb b/docs/source/notebooks/1.2-Categorical_HGF.ipynb
index f4a42850f..06ab288d2 100644
--- a/docs/source/notebooks/1.2-Categorical_HGF.ipynb
+++ b/docs/source/notebooks/1.2-Categorical_HGF.ipynb
@@ -20,7 +20,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.3-CAtegorical_HGF.ipynb)\n",
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.3-CAtegorical_HGF.ipynb)\n",
"\n",
"```{warning}\n",
"The categorical state node and the categorical state-transition nodes are still work in progress. The examples provided here are given for illustration. Things may change or not work until the official publication.\n",
diff --git a/docs/source/notebooks/1.3-Continuous_HGF.ipynb b/docs/source/notebooks/1.3-Continuous_HGF.ipynb
index 84e0def7e..db1e94681 100644
--- a/docs/source/notebooks/1.3-Continuous_HGF.ipynb
+++ b/docs/source/notebooks/1.3-Continuous_HGF.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.3-Continuous_HGF.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.3-Continuous_HGF.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/2-Using_custom_response_functions.ipynb b/docs/source/notebooks/2-Using_custom_response_functions.ipynb
index 71554bcb4..774e43dad 100644
--- a/docs/source/notebooks/2-Using_custom_response_functions.ipynb
+++ b/docs/source/notebooks/2-Using_custom_response_functions.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/2-Using_custom_response_functions.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/2-Using_custom_response_functions.ipynb)"
]
},
{
@@ -542,12 +542,12 @@
"\n",
"Let's now consider that the two vectors of observations $u$ and responses $y$ were obtained from a real participant undergoing a real experiment. In this situation, we assume that this participant internally used a {term}`Perceptual model` and a {term}`Decision rule` that might resemble what we defined previously, and we want to infer what are the most likely values for critical parameters in the model (e.g. the evolution rate $\\omega_2$). To do so, we are going to use our dataset (both $u$ and $y$), and try many models. We are going to fix the values of all HGF parameters to reasonable estimates (here, using the exact same values as in the simulation), except for $\\omega_2$. For this last parameter, we will assume a prior set at $\\mathcal{N}(-2.0, 2.0)$. The idea is that we want to sample many $\\omega_2$ values from this distribution and see if the model is performing better with some values.\n",
"\n",
- "But here, we need a clear definition of what this means *to perform better* for a given model. And this is exactly what a {term}`Response model` does, it is a way for us to evaluate how likely the behaviours $y$ for a given {term}`Perceptual model`, and assuming that the participants use this specific {term}`Decision rule`. In [pyhgf](https://github.com/ilabcode/pyhgf), this step is performed by creating the corresponding {term}`Response function`, which is the Python function that will return the surprise $S$ of getting these behaviours from the participant under this decision rule.\n",
+ "But here, we need a clear definition of what this means *to perform better* for a given model. And this is exactly what a {term}`Response model` does, it is a way for us to evaluate how likely the behaviours $y$ for a given {term}`Perceptual model`, and assuming that the participants use this specific {term}`Decision rule`. In [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), this step is performed by creating the corresponding {term}`Response function`, which is the Python function that will return the surprise $S$ of getting these behaviours from the participant under this decision rule.\n",
"\n",
"````{hint} What is a *response function*?\n",
"Most of the work around HGFs consists in creating and adapting {term}`Response model` to work with a given experimental design. There is no limit in terms of what can be used as a {term}`Response model`, provided that the {term}`Perceptual model` and the {term}`Decision rule` are clearly defined.\n",
"\n",
- "In [pyhgf](https://github.com/ilabcode/pyhgf), the {term}`Perceptual model` is the probabilistic network created with the main {py:class}`pyhgf.model.HGF` and {py:class}`pyhgf.distribution.HGFDistribution` classes. The {term}`Response model` is something that is implicitly defined when we create the {term}`Response function`, a Python function that computes the negative of the log-likelihood of the actions given the perceptual model. This {term}`Response function` can be passed as an argument to the main classes using the keywords arguments `response_function`, `response_function_inputs` and `response_function_parameters`. The `response_function` can be any callable that returns the surprise $S$ of observing action $y$ given this model, and the {term}`Decision rule`. The `response_function_inputs` are the additional data to the response function (optional) while `response_function_parameters` are the additional parameters (optional). The `response_function_inputs` is where the actions $y$ should be provided.\n",
+ "In [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), the {term}`Perceptual model` is the probabilistic network created with the main {py:class}`pyhgf.model.HGF` and {py:class}`pyhgf.distribution.HGFDistribution` classes. The {term}`Response model` is something that is implicitly defined when we create the {term}`Response function`, a Python function that computes the negative of the log-likelihood of the actions given the perceptual model. This {term}`Response function` can be passed as an argument to the main classes using the keywords arguments `response_function`, `response_function_inputs` and `response_function_parameters`. The `response_function` can be any callable that returns the surprise $S$ of observing action $y$ given this model, and the {term}`Decision rule`. The `response_function_inputs` are the additional data to the response function (optional) while `response_function_parameters` are the additional parameters (optional). The `response_function_inputs` is where the actions $y$ should be provided.\n",
"\n",
"```{important}\n",
"A *response function* should not return the actions given perceptual inputs $y | u$ (this is what the {term}`Decision rule` does), but the [surprise](https://en.wikipedia.org/wiki/Information_content) $S$ associated with the observation of actions given the perceptual inputs $S(y | u)$, which is defined by:\n",
@@ -559,7 +559,7 @@
"$$\n",
"```\n",
"\n",
- "If you are already familiar with using HGFs in the Julia equivalent of [pyhgf](https://github.com/ilabcode/pyhgf), you probably noted that the toolbox is split into a **perceptual** package [HierarchicalGaussianFiltering.jl](https://github.com/ilabcode/HierarchicalGaussianFiltering.jl) and a **response** package [ActionModels.jl](https://github.com/ilabcode/ActionModels.jl). This was made to make the difference between the two parts of the HGF clear and be explicit that you can use a perceptual model without any action model. In [pyhgf](https://github.com/ilabcode/pyhgf) however, everything happens in the same package, the response function is merely an optional, additional argument that can be passed to describe how surprise is computed.\n",
+ "If you are already familiar with using HGFs in the Julia equivalent of [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), you probably noted that the toolbox is split into a **perceptual** package [HierarchicalGaussianFiltering.jl](https://github.com/ComputationalPsychiatry/HierarchicalGaussianFiltering.jl) and a **response** package [ActionModels.jl](https://github.com/ComputationalPsychiatry/ActionModels.jl). This was made to make the difference between the two parts of the HGF clear and be explicit that you can use a perceptual model without any action model. In [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) however, everything happens in the same package, the response function is merely an optional, additional argument that can be passed to describe how surprise is computed.\n",
"````"
]
},
@@ -637,7 +637,7 @@
},
"source": [
"```{note}\n",
- "Note here that our {term}`Response function` has a structure that is the standard way to write response functions in [pyhgf](https://github.com/ilabcode/pyhgf), that is with two input arguments:\n",
+ "Note here that our {term}`Response function` has a structure that is the standard way to write response functions in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), that is with two input arguments:\n",
"- the HGF model on which the response function applies (i.e. the {term}`Perceptual model`)\n",
"- the additional parameters provided to the response function. This can include additional parameters that can be part of the equation of the model, or the input data used by this model. We then provide the `response` vector ($y$) here.\n",
"\n",
@@ -1014,7 +1014,7 @@
"```{glossary}\n",
"\n",
"Perceptual model\n",
- " The perceptual model of a Hierarchical Gaussian Filter traditionally refers to the branch receiving observations $u$ about states of the world and that performs the updating of beliefs about these states. By generalisation, the perceptual model is any probabilistic network that can be created in [pyhgf](https://github.com/ilabcode/pyhgf), receiving an arbitrary number of inputs. An HGF that only consists of a perceptual model will act as a Bayesian filter.\n",
+ " The perceptual model of a Hierarchical Gaussian Filter traditionally refers to the branch receiving observations $u$ about states of the world and that performs the updating of beliefs about these states. By generalisation, the perceptual model is any probabilistic network that can be created in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf), receiving an arbitrary number of inputs. An HGF that only consists of a perceptual model will act as a Bayesian filter.\n",
"\n",
"Response model\n",
" The response model of a Hierarchical Gaussian filter refers to the branch that uses the beliefs about the state of the world to generate actions using the {term}`Decision rule`. This branch is also sometimes referred to as the **decision model** or the **observation model**, depending on the fields. Critically, this part of the model can return the surprise ($-\\log[Pr(x)]$) associated with the observations (here, the observations include the inputs $u$ of the probabilistic network, but will also include the responses of the participant $y$ if there are some).\n",
@@ -1023,7 +1023,7 @@
" The decision rule is a function stating how the agent selects among all possible actions, given the state of the beliefs in the perceptual model, and optionally additional parameters. Programmatically, this is a Python function taking a perceptual model as input (i.e. an instance of the HGF class), and returning a sequence of actions. This can be used for simulation. The decision rule should be clearly defined in order to write the {term}`Response function`.\n",
"\n",
"Response function\n",
- " The response function is a term that we use specifically for this package ([pyhgf](https://github.com/ilabcode/pyhgf)). It refers to the Python function that, using a given HGF model and optional parameter, returns the surprise associated with the observed actions.\n",
+ " The response function is a term that we use specifically for this package ([pyhgf](https://github.com/ComputationalPsychiatry/pyhgf)). It refers to the Python function that, using a given HGF model and optional parameter, returns the surprise associated with the observed actions.\n",
"\n",
"```"
]
diff --git a/docs/source/notebooks/3-Multilevel_HGF.ipynb b/docs/source/notebooks/3-Multilevel_HGF.ipynb
index 691aea4eb..c7abd2603 100644
--- a/docs/source/notebooks/3-Multilevel_HGF.ipynb
+++ b/docs/source/notebooks/3-Multilevel_HGF.ipynb
@@ -32,7 +32,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/3-Multilevel_HGF.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/3-Multilevel_HGF.ipynb)"
]
},
{
@@ -137,7 +137,7 @@
"Luckily, we already have all the components in place to do that. We already used Bayesian networks in the previous sections when we were inferring the distribution of some parameters. Here, we only had one agent (i.e. one participant), and therefore did not need any hyperprior. We need to extend this approach a bit, and explicitly state that we want to fit many models (participants) simultaneously, and draw the values of some parameters from a hyper-prior (i.e. the group-level distribution).\n",
"\n",
"But before we move forward, maybe it is worth clarifying some of the terminology we use, especially as, starting from now, many things are called **networks** but are pointing to different parts of the workflow. We can indeed distinguish two kinds:\n",
- "1. The predictive coding neural networks. This is the kind of network that [pyhgf](https://github.com/ilabcode/pyhgf) is designed to handle (see {ref}`probabilistic_networks`). Every HGF model is an instance of such a network.\n",
+ "1. The predictive coding neural networks. This is the kind of network that [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) is designed to handle (see {ref}`probabilistic_networks`). Every HGF model is an instance of such a network.\n",
"2. The Bayesian (multilevel) network is the computational graph that is created with tools like [pymc](https://www.pymc.io/welcome.html). This graph will represent the dependencies between our variables and the way they are transformed.\n",
"\n",
"In this notebook, we are going to create the second type of network and incorporate many networks of the first type in it as custom distribution."
diff --git a/docs/source/notebooks/4-Parameter_recovery.ipynb b/docs/source/notebooks/4-Parameter_recovery.ipynb
index 1120471f0..f019c30d4 100644
--- a/docs/source/notebooks/4-Parameter_recovery.ipynb
+++ b/docs/source/notebooks/4-Parameter_recovery.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/4-Parameter_recovery.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/4-Parameter_recovery.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/5-Non_linear_value_coupling.ipynb b/docs/source/notebooks/5-Non_linear_value_coupling.ipynb
index 1daa5cdf7..cb78a2d8c 100644
--- a/docs/source/notebooks/5-Non_linear_value_coupling.ipynb
+++ b/docs/source/notebooks/5-Non_linear_value_coupling.ipynb
@@ -12,7 +12,7 @@
"cell_type": "markdown",
"metadata": {},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/1.4-Continuous_HGF_non_linear_value_coupling.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/1.4-Continuous_HGF_non_linear_value_coupling.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb b/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb
index 8ef988a6d..8078c3a78 100644
--- a/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb
+++ b/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Example_1_Heart_rate_variability.ipynb)"
]
},
{
@@ -106,7 +106,7 @@
"id": "48020213-7f26-4201-b4b9-f072231bc225",
"metadata": {},
"source": [
- "The nodalized version of the Hierarchical Gaussian Filter that is implemented in [pyhgf](https://github.com/ilabcode/pyhgf) opens the possibility to create filters with multiple inputs. Here, we illustrate how we can use this feature to create an agent that is filtering their physiological signals in real-time. We use a two-level Hierarchical Gaussian Filter to predict the dynamics of the instantaneous heart rate (the RR interval measured at each heartbeat). We then extract the trajectory of surprise at each predictive node to relate it with the cognitive task performed by the participant while the signal is being recorded."
+ "The nodalized version of the Hierarchical Gaussian Filter that is implemented in [pyhgf](https://github.com/ComputationalPsychiatry/pyhgf) opens the possibility to create filters with multiple inputs. Here, we illustrate how we can use this feature to create an agent that is filtering their physiological signals in real-time. We use a two-level Hierarchical Gaussian Filter to predict the dynamics of the instantaneous heart rate (the RR interval measured at each heartbeat). We then extract the trajectory of surprise at each predictive node to relate it with the cognitive task performed by the participant while the signal is being recorded."
]
},
{
diff --git a/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb b/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb
index b11635c5a..5bbf09532 100644
--- a/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb
+++ b/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Example_2_Input_node_volatility_coupling.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb b/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb
index 4ce529bfa..955bde00a 100644
--- a/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb
+++ b/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Example_3_Multi_armed_bandit.ipynb)"
]
},
{
diff --git a/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb b/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb
index 03c230d81..bc126d259 100644
--- a/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb
+++ b/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Exercise_1_Introduction_to_the_generalised_hierarchical_gaussian_filter.ipynb)"
]
},
{
@@ -137,7 +137,7 @@
"x_1^{(k)} \\sim \\mathcal{N}(x_1^{(k-1)}, \\sigma^2)\n",
"$$\n",
"\n",
- "where $\\sigma^2$ is the fixed variance of the distribution. You can find more details on this as well as some code to get started with the simulations in the first tutorial on the [PyHGF documentation](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#).\n",
+ "where $\\sigma^2$ is the fixed variance of the distribution. You can find more details on this as well as some code to get started with the simulations in the first tutorial on the [PyHGF documentation](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#).\n",
"\n",
"```{exercise}\n",
":label: exercise1.1\n",
@@ -183,7 +183,7 @@
"$$\n",
"\n",
"```{hint}\n",
- "This generative process reads top-down: the node higher in the hierarchy ($x_2$) generates new values and passes them to the child nodes. We can also control how much the node $x_1$ is influenced by its previous (see examples [here](https://ilabcode.github.io/pyhgf/notebooks/0.1-Theory.html#value-coupling)). To keep things simple in this exercise, we will assume that $x_1$ is fully influenced by its previous value, as noted in the equation, therefore $x_2$ can be seen as a *drift parent*.\n",
+ "This generative process reads top-down: the node higher in the hierarchy ($x_2$) generates new values and passes them to the child nodes. We can also control how much the node $x_1$ is influenced by its previous (see examples [here](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.1-Theory.html#value-coupling)). To keep things simple in this exercise, we will assume that $x_1$ is fully influenced by its previous value, as noted in the equation, therefore $x_2$ can be seen as a *drift parent*.\n",
"```\n",
"\n",
"Let's define a time series `x_2` that represent the evolution of the states of the parent node:"
@@ -373,11 +373,11 @@
"source": [
"Building on these principles, we can create networks of arbitrary size and shape made of probabilistic nodes (nodes that represent the evolution of a Gaussian Random Walk) that can be connected with each other through **value** or **volatility** coupling. We have covered these points in the previous exercise using a simplified network made of two nodes.\n",
"\n",
- "[PyHGF](https://ilabcode.github.io/pyhgf/index.html) adds most of its values in that it allows the creation and manipulation of large and complex networks easily, and it automates the transmission of values between nodes. You can refer to [the second part of the tutorial documentation](https://ilabcode.github.io/pyhgf/notebooks/0.2-Creating_networks.html) which covers the manipulation of networks for more details. \n",
+ "[PyHGF](https://computationalpsychiatry.github.io/pyhgf/index.html) adds most of its values in that it allows the creation and manipulation of large and complex networks easily, and it automates the transmission of values between nodes. You can refer to [the second part of the tutorial documentation](https://computationalpsychiatry.github.io/pyhgf/notebooks/0.2-Creating_networks.html) which covers the manipulation of networks for more details. \n",
"\n",
"But this is not the only thing it does. So far we have simulated observations from the generative model, going for the leaves to the root of the network. This describes how we expect the environment to behave, but not how an agent would learn from it. To do so we need to invert this model: we want to provide the observations, and let the network update the nodes accordingly, so the beliefs always reflect the current state of the environment. This process requires propagating precision prediction errors from the root to the leaves of the network, and this is where most of PyHGF's dark magic is most useful.\n",
"\n",
- "```{figure} https://raw.githubusercontent.com/ilabcode/pyhgf/master/docs/source/images/graph_network.svg\n",
+ "```{figure} https://raw.githubusercontent.com/ComputationalPsychiatry/pyhgf/master/docs/source/images/graph_network.svg\n",
"---\n",
"name: networks\n",
"---\n",
@@ -760,7 +760,7 @@
],
"source": [
"aarhus_weather_df = pd.read_csv(\n",
- " \"https://raw.githubusercontent.com/ilabcode/hgf-data/main/datasets/weather.csv\"\n",
+ " \"https://raw.githubusercontent.com/ComputationalPsychiatry/hgf-data/main/datasets/weather.csv\"\n",
")\n",
"aarhus_weather_df.head()"
]
@@ -1076,7 +1076,7 @@
"````{solution} exercise1.5\n",
":label: solution-exercise1.5\n",
"\n",
- "This method return the Gaussian surprise at each time point, which are then summed. The sum of the Gaussian surprise reflect the performances of the model at predicting the next value, larger values pointing to a model more surprise by its inputs, therefore poor performances. The surprise is define as $s = -log(p)$, this is thus the negative of the log probability function. Log probability functions are commonly used by sampling algorithm, it is thus straigthforwars to sample a model parameters when this function is available. There are an infinity of response possible functions - just like there is an infinity of possible networks - for more details on how to use tem, you can refer to the [tutorial on custom response models](https://ilabcode.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html).\n",
+ "This method return the Gaussian surprise at each time point, which are then summed. The sum of the Gaussian surprise reflect the performances of the model at predicting the next value, larger values pointing to a model more surprise by its inputs, therefore poor performances. The surprise is define as $s = -log(p)$, this is thus the negative of the log probability function. Log probability functions are commonly used by sampling algorithm, it is thus straigthforwars to sample a model parameters when this function is available. There are an infinity of response possible functions - just like there is an infinity of possible networks - for more details on how to use tem, you can refer to the [tutorial on custom response models](https://computationalpsychiatry.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html).\n",
"\n",
"````"
]
diff --git a/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb b/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
index 7a1b0f4c1..0dc3ce8c6 100644
--- a/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
+++ b/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb
@@ -26,7 +26,7 @@
"tags": []
},
"source": [
- "[](https://colab.research.google.com/github/ilabcode/pyhgf/blob/master/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb)"
+ "[](https://colab.research.google.com/github/ComputationalPsychiatry/pyhgf/blob/master/docs/source/notebooks/Exercise_2_Bayesian_reinforcement_learning.ipynb)"
]
},
{
@@ -1942,9 +1942,9 @@
"\n",
"All these sections should give you a solid understanding of the model and how to use it in context. If you want to apply this to your dataset, we recommend exploring the tutorial section of the documentation:\n",
"\n",
- "- [Using custom response models](https://ilabcode.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html) will explore the creation of custom models for behaviours that can match your experimental design and theory.\n",
- "- [Multilevel modelling](https://ilabcode.github.io/pyhgf/notebooks/3-Multilevel_HGF.html) will discuss modelling at the group/condition level in multilevel Bayesian networks.\n",
- "- [Parameter recovery](https://ilabcode.github.io/pyhgf/notebooks/4-Parameter_recovery.html) explains how to simulate a dataset and perform parameter recovery, as a prior validation of your models."
+ "- [Using custom response models](https://computationalpsychiatry.github.io/pyhgf/notebooks/2-Using_custom_response_functions.html) will explore the creation of custom models for behaviours that can match your experimental design and theory.\n",
+ "- [Multilevel modelling](https://computationalpsychiatry.github.io/pyhgf/notebooks/3-Multilevel_HGF.html) will discuss modelling at the group/condition level in multilevel Bayesian networks.\n",
+ "- [Parameter recovery](https://computationalpsychiatry.github.io/pyhgf/notebooks/4-Parameter_recovery.html) explains how to simulate a dataset and perform parameter recovery, as a prior validation of your models."
]
},
{
diff --git a/pyproject.toml b/pyproject.toml
index 678d407fd..d9a4246d3 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -4,8 +4,8 @@ description = "Dynamic neural networks for predictive coding"
authors = ["Nicolas Legrand